MEG Biowulf Setup: Difference between revisions
Jump to navigation
Jump to search
Content deleted Content added
| (6 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
= Quick tutorial to generate most of the needed inputs for data analysis = |
|||
== General Setup == |
== General Setup == |
||
=== Mounting local computer to biowulf === |
=== Mounting local computer to biowulf === |
||
https://hpc.nih.gov/docs/hpcdrive.html |
https://hpc.nih.gov/docs/hpcdrive.html |
||
== Configuring your bash shell environment == |
== Configuring your bash shell environment == |
||
If editing your bashrc -- open two terminals in biowulf. If you misconfigure your .bashrc, you |
If editing your bashrc -- open two terminals in biowulf. '''If you misconfigure your .bashrc, you might not be able to log into biowulf'''. Having two terminals open allows you to fix anything that errors out. |
||
=== Edit .bashrc file in your home drive === |
=== Edit .bashrc file in your home drive === |
||
Aliases allow you to configure commands for easy use. <br> |
|||
If you have a data directory that you use, you may want to add '''cddat="cd /data/MYDATADIR/blah/blah/data"''' |
|||
umask 002 #Gives automatic group permissions to every file you create -- very very helpful for working with your team |
umask 002 #Gives automatic group permissions to every file you create -- very very helpful for working with your team |
||
#Add modules bin path to access the MEG modules |
|||
PATH=/data/MEGmodules/bin:$PATH |
|||
## Set up some aliases, so you don't have to type these out |
## Set up some aliases, so you don't have to type these out |
||
| Line 16: | Line 14: | ||
alias sinteractive_medium='sinteractive --mem=16G --cpus-per-task=12 --gres=lscratch:100' |
alias sinteractive_medium='sinteractive --mem=16G --cpus-per-task=12 --gres=lscratch:100' |
||
alias sinteractive_large='sinteractive --mem=24G --cpus-per-task=32 --gres=lscratch:150' |
alias sinteractive_large='sinteractive --mem=24G --cpus-per-task=32 --gres=lscratch:150' |
||
=== Edit .bash_profile in your home drive === |
|||
#Set your default group (normally your default is your userID - which isn't helpful for your group |
|||
#Type `groups` to see which groups you are part of |
|||
newgrp <<YOUR GROUP ID>> |
|||
=== To Access Additional MEG modules === |
=== To Access Additional MEG modules === |
||
| Line 31: | Line 24: | ||
spyder |
spyder |
||
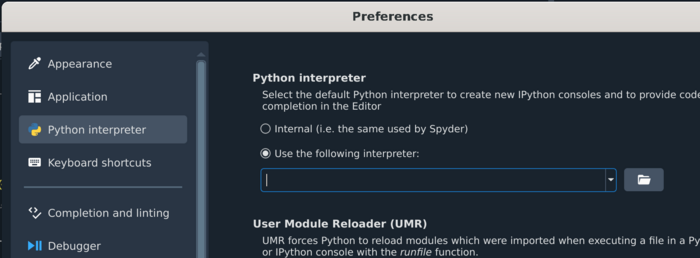
Click on the wrench icon <br> |
|||
| ⚫ | |||
[[File:Spyder_config_button.png | 700px]] <br><br> |
|||
== General Processing Pipeline == |
|||
=== First write out the appropriate events in the markerfile === |
|||
TBD - fill in items here |
|||
=== Convert Data to BIDS === |
|||
module load mne |
|||
make_meg_bids.py -h #Then fill in the required items |
|||
== Processing the MRI related components == |
|||
#Generate Bounary Element Model / Transform / Forward model for source localization. Check the help, there is a volume vs surface flag |
|||
#This will write out the swarmfile (which can be editted) -- and prints out the command to launch the swarm job |
|||
megcore_prep_mri_bids.py -gen_swarmfile -bids_root <<BIDS_ROOT>> -swarm_fname <<SWARM_FNAME>> -subject <<SUBJECT>> -run <<RUN>> -session <<SESSION>> -task <<TASK>> |
|||
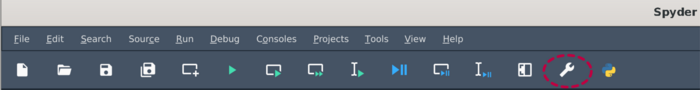
| ⚫ | |||
== Parsing Logfiles == |
|||
[[File:Preferences panel spyder.png | 700px]] <br> |
|||
Presentation Files: |
|||
https://github.com/gjcooper/prespy |
|||
Latest revision as of 15:17, 24 September 2025
General Setup
Mounting local computer to biowulf
https://hpc.nih.gov/docs/hpcdrive.html
Configuring your bash shell environment
If editing your bashrc -- open two terminals in biowulf. If you misconfigure your .bashrc, you might not be able to log into biowulf. Having two terminals open allows you to fix anything that errors out.
Edit .bashrc file in your home drive
Aliases allow you to configure commands for easy use.
If you have a data directory that you use, you may want to add cddat="cd /data/MYDATADIR/blah/blah/data"
umask 002 #Gives automatic group permissions to every file you create -- very very helpful for working with your team ## Set up some aliases, so you don't have to type these out alias sinteractive_small='sinteractive --mem=8G --cpus-per-task=4 --gres=lscratch:30' alias sinteractive_medium='sinteractive --mem=16G --cpus-per-task=12 --gres=lscratch:100' alias sinteractive_large='sinteractive --mem=24G --cpus-per-task=32 --gres=lscratch:150'
To Access Additional MEG modules
#Add the following line to your ${HOME}/.bashrc
module use --append /data/MEGmodules/modulefiles
Setting up spyder on biowulf
module purge module load python/3.10 spyder
Add the following line to Use the following interpreter: /vf/users/MEGmodules/modules/mne1.5.1dev/bin/python3.11