MEG Biowulf Setup
Jump to navigation
Jump to search
General Setup
Mounting local computer to biowulf
https://hpc.nih.gov/docs/hpcdrive.html
Configuring your bash shell environment
If editing your bashrc -- open two terminals in biowulf. If you misconfigure your .bashrc, you might not be able to log into biowulf. Having two terminals open allows you to fix anything that errors out.
Edit .bashrc file in your home drive
Aliases allow you to configure commands for easy use.
If you have a data directory that you use, you may want to add cddat="cd /data/MYDATADIR/blah/blah/data"
umask 002 #Gives automatic group permissions to every file you create -- very very helpful for working with your team ## Set up some aliases, so you don't have to type these out alias sinteractive_small='sinteractive --mem=8G --cpus-per-task=4 --gres=lscratch:30' alias sinteractive_medium='sinteractive --mem=16G --cpus-per-task=12 --gres=lscratch:100' alias sinteractive_large='sinteractive --mem=24G --cpus-per-task=32 --gres=lscratch:150'
To Access Additional MEG modules
#Add the following line to your ${HOME}/.bashrc
module use --append /data/MEGmodules/modulefiles
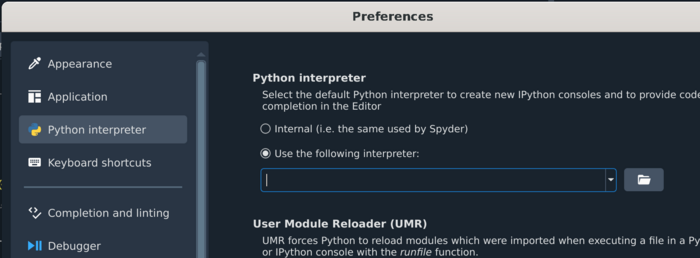
Setting up spyder on biowulf
module purge module load python/3.10 spyder
Add the following line to Use the following interpreter: /vf/users/MEGmodules/modules/mne1.5.1dev/bin/python3.11