BIDS GUIs: Difference between revisions
| (42 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
==What is BIDS== |
==What is BIDS== |
||
BIDS is a standard specification for neuroimaging/physiology data. This currently includes at least: MRI, fMRI, DTI, EEG, MEG, fNIRS (and possibly ECOG/sEEG). BIDS typically describes how RAW data is organized - and processed data is located in the bids_dir/'''derivatives'''/{AnalysisPackage}/{SUBJECT}/... The main advantage is that common code can be generated to process data organized in a standard format. Therefore, you should be able to import the bids data into any number of neurophysiological packages (MNE, Brainstorm, SPM, Fieldtrip, ...). Additionally, standardized processing packages known as BIDS apps can be used to process the data in the same way as long as the data is organized in BIDS. |
|||
https://bids-specification.readthedocs.io/en/stable/ |
|||
https://bids-specification.readthedocs.io/en/stable/modality-specific-files/magnetoencephalography.html <br> |
|||
BIDS is a standard specification for neuroimaging/physiology data. This currently includes at least: MRI, fMRI, DTI, EEG, MEG, fNIRS (and possibly ECOG/sEEG). BIDS typically describes how RAW data is organized - and processed data is located in the bids_dir/derivatives/{AnalysisPackage}/{SUBJECT}/... The main advantage is that common code can be generated to process data organized in a standard format. Therefore, you should be able to import the bids data into any number of neurophysiological packages (MNE, Brainstorm, SPM, Fieldtrip, ...). Additionally, standardized processing packages known as BIDS apps can be used to process the data in the same way as long as the data is organized in BIDS. |
|||
https://megcore.nih.gov/index.php/Mne_bids_pipeline <br> |
|||
==Biowulf Setup== |
|||
https://megcore.nih.gov/index.php/MEG_Biowulf_Setup |
|||
==On Biowulf== |
==On Biowulf== |
||
Log into biowulf using NoMachine. Setup instructions here: https://hpc.nih.gov/docs/nx.html <br> |
|||
Start a compute node from the terminal |
|||
NOTE: Remember to change the resolution to be close to your monitor (top right corner) <br> |
|||
sinteractive --mem=12G --cpus-per-task=4 --gres=lscratch:50 #adjust mem and cpus accordingly |
|||
On biowulf open a terminal and start a compute node using the commands below |
|||
sinteractive --mem=24G --cpus-per-task=4 --gres=lscratch:50 #adjust mem and cpus accordingly |
|||
module use --append /data/MEGmodules/modulefiles |
module use --append /data/MEGmodules/modulefiles |
||
module load mne/dev1.5.1 |
module load mne/dev1.5.1 |
||
==Prep Data for BIDS== |
|||
Add event coding to the datasets before converting to bids <br> |
|||
Launch the GUI: (will eventually add this to the path -- currently needs to call with the full path) |
|||
/vf/users/MEGmodules/modules/mne1.5.1dev_extras/nih_to_mne/nih2mne/GUI/trigger_code_gui.py |
|||
Open a dataset: <br> |
|||
This will read all trigger lines (analog and digital) and populate lines <br> |
|||
The triggers can be evaluated on upgoing or downgoing portions of the trigger squarewave <br> |
|||
[[File:Trigger_gui_initial_loading.png | 900px]] <br><br> |
|||
Name all of the appropriate trigger lines (digital and analogue codes) <br> |
|||
Hitting the '''Update Event Names''' button to access the derivative event menu. You can now make events from other events. <br> |
|||
'''DO NOT EDIT THE TOP PANEL AFTER HITTING THIS BUTTON''' <br> |
|||
[[File:Trigger_gui_initial_entry.png | 900px]] <br> |
|||
The derivative event panel allows you to make new events from other events. <br> |
|||
Select a lead event from the dropdown (the menu will display any events that have been established above the current line) <br> |
|||
Select a lag event from the dropdown <br> |
|||
Click the checkbox next to the event that should establish the timing. <br> |
|||
Enter the window to search from the lead event (this can include negative numbers in the initial entry) <br> |
|||
Enter the name that the new derivative event will be called. |
|||
Click the '''ADD''' button. This will add a new line and add this event to the dropdown list and the keep events panel at the bottom <br> |
|||
[[File:Trigger_gui_parsemarks_demo.png | 700px]] <br> |
|||
Anything not named cannot be used below to parse two input codes or to save out to the MarkerFile.mrk event code file <br> |
|||
Once finished coding the events - the bottom panel determines which events are written to the Markerfile - intermediary events not selected will be ignored<br> |
|||
D-prime coding is often used (Hit, Miss, False Alarm, Correct Rejection) |
|||
Click '''Write Parser File''' to generate a script that can be used for all subjects under this protocol <br> |
|||
[[File:Event_processing_generated_script.png | 700px]]<br> |
|||
<br> |
|||
Using the processing script: <br> |
|||
# Set the permissions of the generated script to excecutable (Only needs to be done once) |
|||
chmod +x trig_processing.sh #Use whatever name you have saved out the file as |
|||
# Use the processing script to add the event coding into your dataset - This can be used on any data acquired with this protocol (and same trigger data lines) |
|||
trig_processing.sh <<DATAFILENAME.ds>> |
|||
After confirming that the processing script works on your dataset - it is beneficial to add this to your github repository for data reproducability. |
|||
You can manually tweak the script as well -- it is just using dataframes to process the event coding. For example a separate logfile that has been been processed into a csv can be read into pandas (pd.read_csv) and appended to the larger dataframe. Then events from the logfile can be used to generate derivative events using the parse_marks command. |
|||
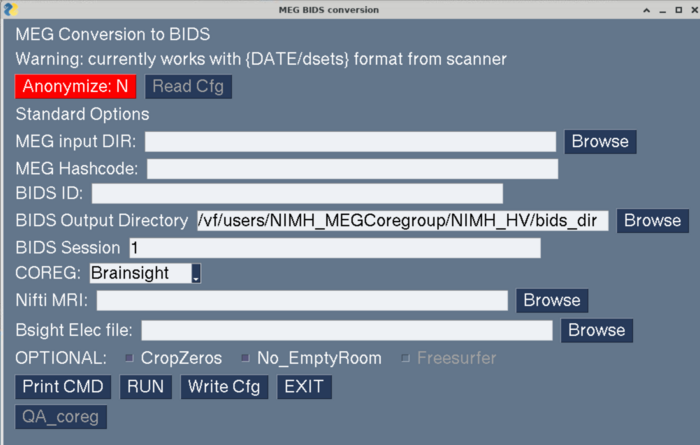
==Create BIDS Dataset Using GUI== |
|||
https://megcore.nih.gov/MEG/BIDS_gui_080224_anon.pdf |
|||
make_meg_bids_gui.py |
|||
[[File:Make_meg_bids_gui_startup.png | 700px]] |
|||
=QA GUI Overview= |
|||
Launch the QA gui |
|||
bids_qa_gui.py -bids_root <<PATH to BIDS dir>> |
bids_qa_gui.py -bids_root <<PATH to BIDS dir>> |
||
If there are issues launch the gui from the BIDS directory |
|||
=GUI Overview= |
|||
cd <<PATH to BIDS DIR>> |
|||
bids_qa_gui.py -bids_root $(pwd) |
|||
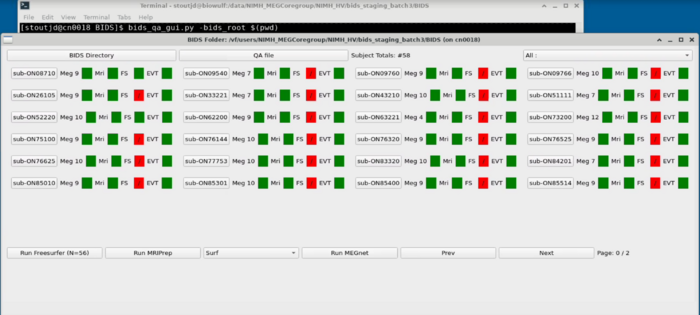
==Project Panel== |
==Project Panel== |
||
[[File:Project_panel_view.png]] |
[[File:Project_panel_view.png | 700px]] |
||
The project panel consists of tiles with each subject ID. <br> |
|||
'''Subject ID Button''': |
|||
* Meg: Number of MEG files |
|||
* Mri: If they have an MRI |
|||
* FS: Has the freesurfer been run and is successful |
|||
* EVT: Events pass QA (Currently this is just green and doesn't QA) |
|||
'''QA File''': <br> |
|||
Set the appropriate number of events in a successful meg dataset<br> |
|||
'''Top Right Dropdown''': <br> |
|||
Filter for specific tasks - this will adjust the color coding on the MEG <br> |
|||
'''Run Freesurfer Button''': <br> |
|||
Submits freesurfer sbatch job to biowulf for each missing subject with an mri <br> |
|||
'''Run MEGnet''': <br> |
|||
Currently a placeholder <br> |
|||
'''Prev/Next''': <br> |
|||
Advance to the next panel if more than a single grid of subjects is available <br> |
|||
==Subject Panel== |
==Subject Panel== |
||
[[File:Gui_subject_panel.png]] |
[[File:Gui_subject_panel.png | 700px]] |
||
==Task Switcher== |
==Task Switcher== |
||
Changing the task - shows the number of epochs in the selected task |
Changing the task - shows the number of epochs in the selected task <br> |
||
[[File:Subject_panel_switch_task_epochs.png]] |
[[File:Subject_panel_switch_task_epochs.png | 700px]] |
||
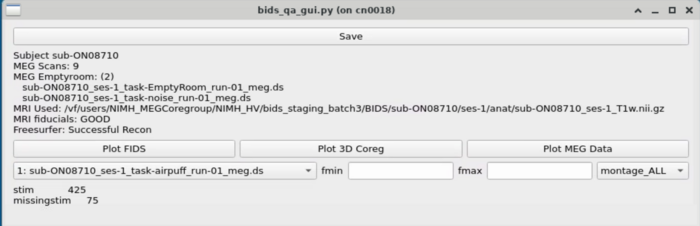
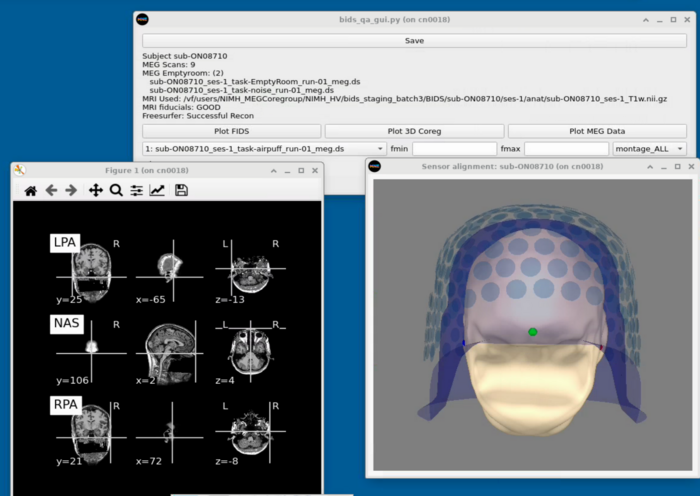
==MRI Viewing== |
==MRI Viewing== |
||
Plot FIDS works on the base MRI and doesn't require any prep. <br> |
|||
[[File:Plot_mri_views.png]] |
|||
To plot the 3D coreg - Freesurfer must be processed on the subject and the MRI preprocessing must have been completed <br> |
|||
[[File:Plot_mri_views.png | 700px ]] |
|||
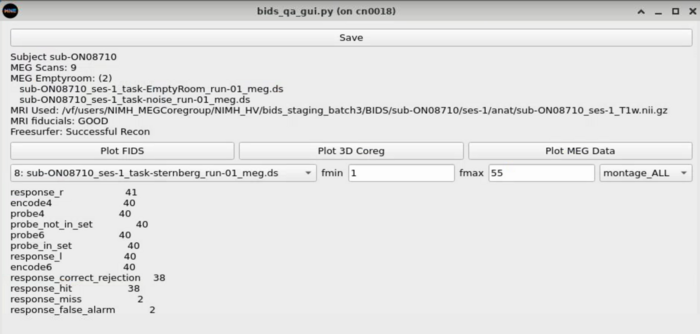
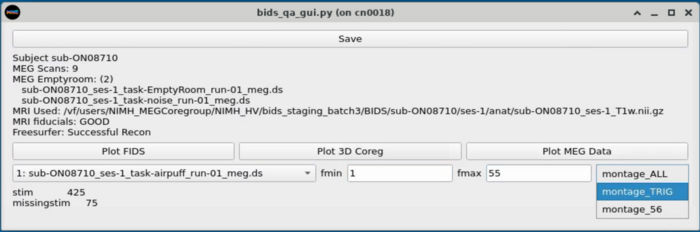
==MEG Plotting== |
==MEG Plotting== |
||
Dropdown with MEG plotting options |
Dropdown with MEG plotting options <br> |
||
[[Meg_plotting_options.png]] |
[[File:Meg_plotting_options.png | 700px]] |
||
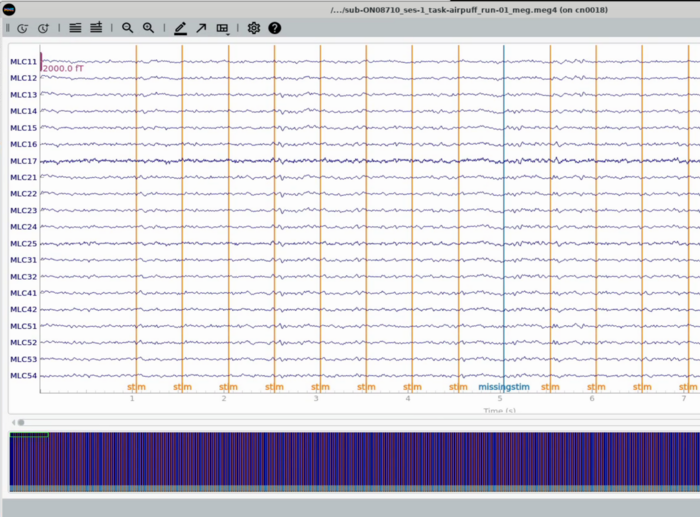
Plot All MEG channels (unorganized view of All channels) <br> |
|||
[[File:Plot_meg_all.png | 700px]] |
|||
Plot whole head - subset of meg channels representing the Left (top half) to Right (bottom half) display <br> |
|||
[[File:Plot_meg_fullview.png | 700px]] |
|||
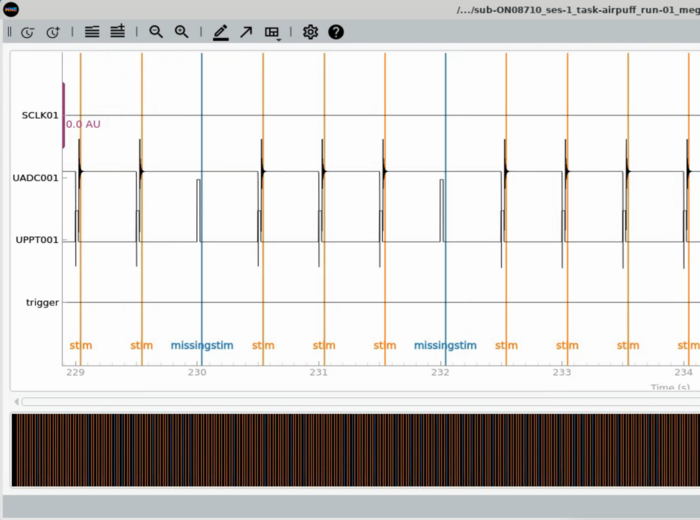
Plot Trigger Channels <br> |
|||
Note the auditory delay added to the triggerline pulse <br> |
|||
[[File:Plot_meg_trigview.png | 700px]] |
|||
Setting BAD Channels and BAD Epochs (Must do WriteBads afterwards) <br> |
|||
Plot All MEG channels (unorganized view) |
|||
1) Plot a dataset (montage56 is preferable) with a wide bandwidth filter fmin=0.5 and fmax=110. The notch should be checked <br> |
|||
[[Plot_meg_all.png]] |
|||
2) BAD Epochs: Click on "Add Description" >> Set the name. The name '''must start''' with '''BAD_''' or it will not be written. <br> |
|||
3) Left click and drag to set a bad time period. <br> |
|||
[[File:Select_bad_epochs.png | 700px]] |
|||
4) Clicking on a channel will set it to grey. This sets the channel to BAD ('''!!Warning it is easy to accidentally do this!!'''). <br> |
|||
Plot Trigger Channels |
|||
5) After closing the plot, you can then click the write Bad Channels and Bad Epochs. <br> |
|||
[[Plot_meg_trigview.png]] |
|||
[[File:Write_bads_button.png | 700px]] <br> |
|||
[[File:Bads_write_confirmation.png | 300px]] |
|||
Latest revision as of 17:50, 31 January 2025
What is BIDS
BIDS is a standard specification for neuroimaging/physiology data. This currently includes at least: MRI, fMRI, DTI, EEG, MEG, fNIRS (and possibly ECOG/sEEG). BIDS typically describes how RAW data is organized - and processed data is located in the bids_dir/derivatives/{AnalysisPackage}/{SUBJECT}/... The main advantage is that common code can be generated to process data organized in a standard format. Therefore, you should be able to import the bids data into any number of neurophysiological packages (MNE, Brainstorm, SPM, Fieldtrip, ...). Additionally, standardized processing packages known as BIDS apps can be used to process the data in the same way as long as the data is organized in BIDS.
https://bids-specification.readthedocs.io/en/stable/modality-specific-files/magnetoencephalography.html
https://megcore.nih.gov/index.php/Mne_bids_pipeline
Biowulf Setup
https://megcore.nih.gov/index.php/MEG_Biowulf_Setup
On Biowulf
Log into biowulf using NoMachine. Setup instructions here: https://hpc.nih.gov/docs/nx.html
NOTE: Remember to change the resolution to be close to your monitor (top right corner)
On biowulf open a terminal and start a compute node using the commands below
sinteractive --mem=24G --cpus-per-task=4 --gres=lscratch:50 #adjust mem and cpus accordingly
module use --append /data/MEGmodules/modulefiles module load mne/dev1.5.1
Prep Data for BIDS
Add event coding to the datasets before converting to bids
Launch the GUI: (will eventually add this to the path -- currently needs to call with the full path)
/vf/users/MEGmodules/modules/mne1.5.1dev_extras/nih_to_mne/nih2mne/GUI/trigger_code_gui.py
Open a dataset:
This will read all trigger lines (analog and digital) and populate lines
The triggers can be evaluated on upgoing or downgoing portions of the trigger squarewave

Name all of the appropriate trigger lines (digital and analogue codes)
Hitting the Update Event Names button to access the derivative event menu. You can now make events from other events.
DO NOT EDIT THE TOP PANEL AFTER HITTING THIS BUTTON

The derivative event panel allows you to make new events from other events.
Select a lead event from the dropdown (the menu will display any events that have been established above the current line)
Select a lag event from the dropdown
Click the checkbox next to the event that should establish the timing.
Enter the window to search from the lead event (this can include negative numbers in the initial entry)
Enter the name that the new derivative event will be called.
Click the ADD button. This will add a new line and add this event to the dropdown list and the keep events panel at the bottom

Anything not named cannot be used below to parse two input codes or to save out to the MarkerFile.mrk event code file
Once finished coding the events - the bottom panel determines which events are written to the Markerfile - intermediary events not selected will be ignored
D-prime coding is often used (Hit, Miss, False Alarm, Correct Rejection)
Click Write Parser File to generate a script that can be used for all subjects under this protocol

Using the processing script:
# Set the permissions of the generated script to excecutable (Only needs to be done once) chmod +x trig_processing.sh #Use whatever name you have saved out the file as
# Use the processing script to add the event coding into your dataset - This can be used on any data acquired with this protocol (and same trigger data lines) trig_processing.sh <<DATAFILENAME.ds>>
After confirming that the processing script works on your dataset - it is beneficial to add this to your github repository for data reproducability. You can manually tweak the script as well -- it is just using dataframes to process the event coding. For example a separate logfile that has been been processed into a csv can be read into pandas (pd.read_csv) and appended to the larger dataframe. Then events from the logfile can be used to generate derivative events using the parse_marks command.
Create BIDS Dataset Using GUI
https://megcore.nih.gov/MEG/BIDS_gui_080224_anon.pdf
make_meg_bids_gui.py
QA GUI Overview
Launch the QA gui
bids_qa_gui.py -bids_root <<PATH to BIDS dir>>
If there are issues launch the gui from the BIDS directory
cd <<PATH to BIDS DIR>> bids_qa_gui.py -bids_root $(pwd)
Project Panel
The project panel consists of tiles with each subject ID.
Subject ID Button:
- Meg: Number of MEG files
- Mri: If they have an MRI
- FS: Has the freesurfer been run and is successful
- EVT: Events pass QA (Currently this is just green and doesn't QA)
QA File:
Set the appropriate number of events in a successful meg dataset
Top Right Dropdown:
Filter for specific tasks - this will adjust the color coding on the MEG
Run Freesurfer Button:
Submits freesurfer sbatch job to biowulf for each missing subject with an mri
Run MEGnet:
Currently a placeholder
Prev/Next:
Advance to the next panel if more than a single grid of subjects is available
Subject Panel
Task Switcher
Changing the task - shows the number of epochs in the selected task
MRI Viewing
Plot FIDS works on the base MRI and doesn't require any prep.
To plot the 3D coreg - Freesurfer must be processed on the subject and the MRI preprocessing must have been completed
MEG Plotting
Dropdown with MEG plotting options
Plot All MEG channels (unorganized view of All channels)
Plot whole head - subset of meg channels representing the Left (top half) to Right (bottom half) display
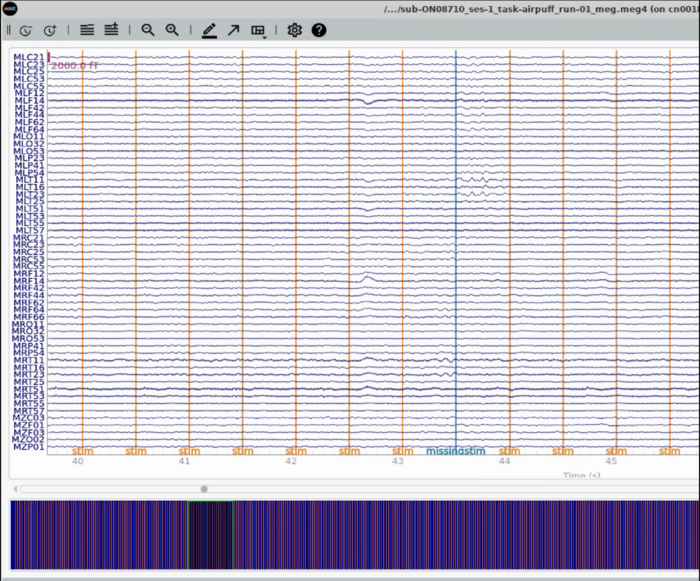
Plot Trigger Channels
Note the auditory delay added to the triggerline pulse
Setting BAD Channels and BAD Epochs (Must do WriteBads afterwards)
1) Plot a dataset (montage56 is preferable) with a wide bandwidth filter fmin=0.5 and fmax=110. The notch should be checked
2) BAD Epochs: Click on "Add Description" >> Set the name. The name must start with BAD_ or it will not be written.
3) Left click and drag to set a bad time period.

4) Clicking on a channel will set it to grey. This sets the channel to BAD (!!Warning it is easy to accidentally do this!!).
5) After closing the plot, you can then click the write Bad Channels and Bad Epochs.