ENIGMA MEG data upload: Difference between revisions
Data anonymization added |
|||
| Line 1: | Line 1: | ||
=Steps prior to data upload= |
|||
| ⚫ | |||
#Anonymize MEG data |
|||
| ⚫ | |||
#Anonymize MRI, deface, confirm correct defacing (options below) |
|||
## Freesurfer: mri_deface : https://surfer.nmr.mgh.harvard.edu/fswiki/mri_deface |
|||
## Pydeface: pydeface : https://github.com/poldracklab/pydeface |
|||
## SPM: spm_deface : https://www.fil.ion.ucl.ac.uk/spm/software/spm12/ |
|||
## Brainstorm : MRI (right click) > Deface Volume : https://neuroimage.usc.edu/brainstorm/Introduction |
|||
## Fieldtrip: ft_defacevolume: https://www.fieldtriptoolbox.org/reference/ft_defacevolume |
|||
## Afni: @afni_refacer_run : https://afni.nimh.nih.gov/pub/dist/doc/htmldoc/tutorials/refacer/refacer_run.html |
|||
## Brainvoyager: : https://download.brainvoyager.com/bv/doc/UsersGuide/GettingStarted/AnonymizationAndDefacing.html |
|||
#Confirm defacing algorithm prior to upload using an MR viewer (many options above) |
|||
| ⚫ | |||
Data to be included in the Enigma MEG mega-analysis and data harmonization procedure will be uploaded using the Globus software. Globus is a secure, fault-tolerant, peer-to-peer software based on GridFTP. Secure control signals are communicated with the Globus website, but data transfer is only performed between endpoints (peer-to-peer). |
Data to be included in the Enigma MEG mega-analysis and data harmonization procedure will be uploaded using the Globus software. Globus is a secure, fault-tolerant, peer-to-peer software based on GridFTP. Secure control signals are communicated with the Globus website, but data transfer is only performed between endpoints (peer-to-peer). |
||
An endpoint per lab will be created on NIH storage and given access credentials via email. Only users within your lab and the NIH MEG Core will be able to view and write data in the provided endpoint. Once you receive an email from globus connecting you to a globus endpoint, you will need to create an account on the globus website associated with your institution. |
An endpoint per lab will be created on NIH storage and given access credentials via email. Only users within your lab and the NIH MEG Core will be able to view and write data in the provided endpoint. Once you receive an email from globus connecting you to a globus endpoint, you will need to create an account on the globus website associated with your institution. |
||
| ⚫ | |||
The globus website will allow you to see the upload endpoint |
The globus website will allow you to see the upload endpoint |
||
https://www.globus.org/ |
https://www.globus.org/ |
||
=Required Software to see local data= |
|||
Globus connect personal is used to browse local data for upload. |
Globus connect personal is used to browse local data for upload. |
||
https://www.globus.org/globus-connect-personal |
https://www.globus.org/globus-connect-personal |
||
=Data upload process= |
|||
| ⚫ | |||
<br> |
|||
| ⚫ | |||
'''*Log into the Globus website to view main menu''' |
'''*Log into the Globus website to view main menu''' |
||
<br> |
<br> |
||
Revision as of 21:06, 25 February 2021
Steps prior to data upload
- Anonymize MEG data
- Anonymize MRI, deface, confirm correct defacing (options below)
- Freesurfer: mri_deface : https://surfer.nmr.mgh.harvard.edu/fswiki/mri_deface
- Pydeface: pydeface : https://github.com/poldracklab/pydeface
- SPM: spm_deface : https://www.fil.ion.ucl.ac.uk/spm/software/spm12/
- Brainstorm : MRI (right click) > Deface Volume : https://neuroimage.usc.edu/brainstorm/Introduction
- Fieldtrip: ft_defacevolume: https://www.fieldtriptoolbox.org/reference/ft_defacevolume
- Afni: @afni_refacer_run : https://afni.nimh.nih.gov/pub/dist/doc/htmldoc/tutorials/refacer/refacer_run.html
- Brainvoyager: : https://download.brainvoyager.com/bv/doc/UsersGuide/GettingStarted/AnonymizationAndDefacing.html
- Confirm defacing algorithm prior to upload using an MR viewer (many options above)
Globus Website
Data to be included in the Enigma MEG mega-analysis and data harmonization procedure will be uploaded using the Globus software. Globus is a secure, fault-tolerant, peer-to-peer software based on GridFTP. Secure control signals are communicated with the Globus website, but data transfer is only performed between endpoints (peer-to-peer).
An endpoint per lab will be created on NIH storage and given access credentials via email. Only users within your lab and the NIH MEG Core will be able to view and write data in the provided endpoint. Once you receive an email from globus connecting you to a globus endpoint, you will need to create an account on the globus website associated with your institution.
The globus website will allow you to see the upload endpoint https://www.globus.org/
Required Software to see local data
Globus connect personal is used to browse local data for upload. https://www.globus.org/globus-connect-personal
Data upload process
---DATA MUST BE ANONYMIZED BEFORE UPLOAD!---
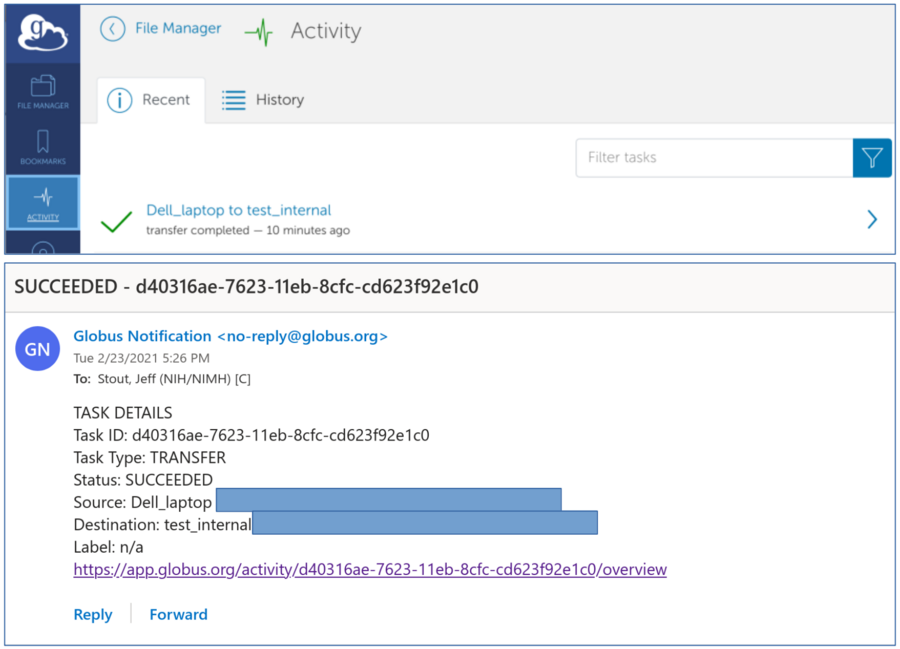
*Log into the Globus website to view main menu
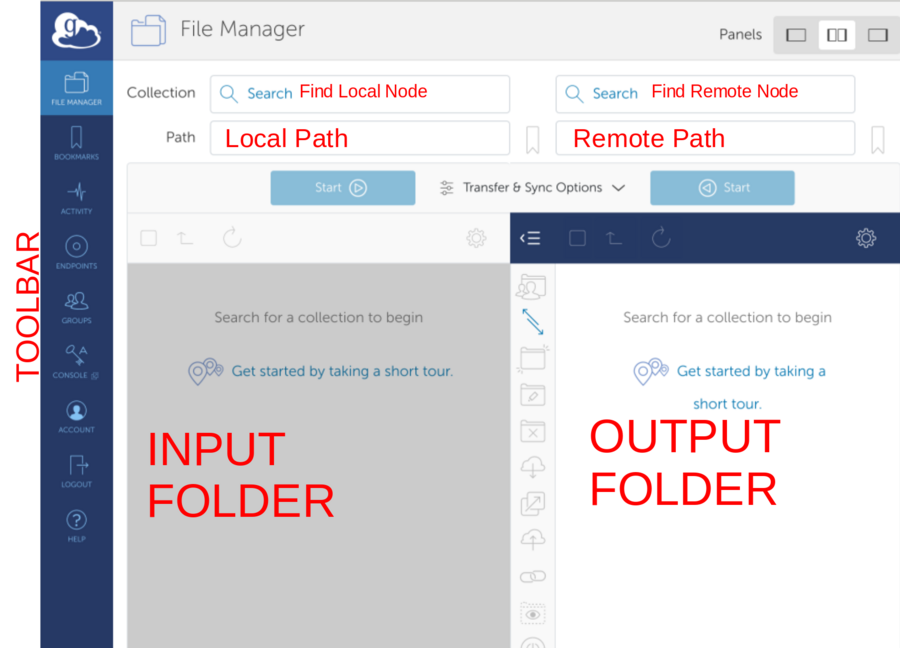
*Find remote endpoint - Click the search field
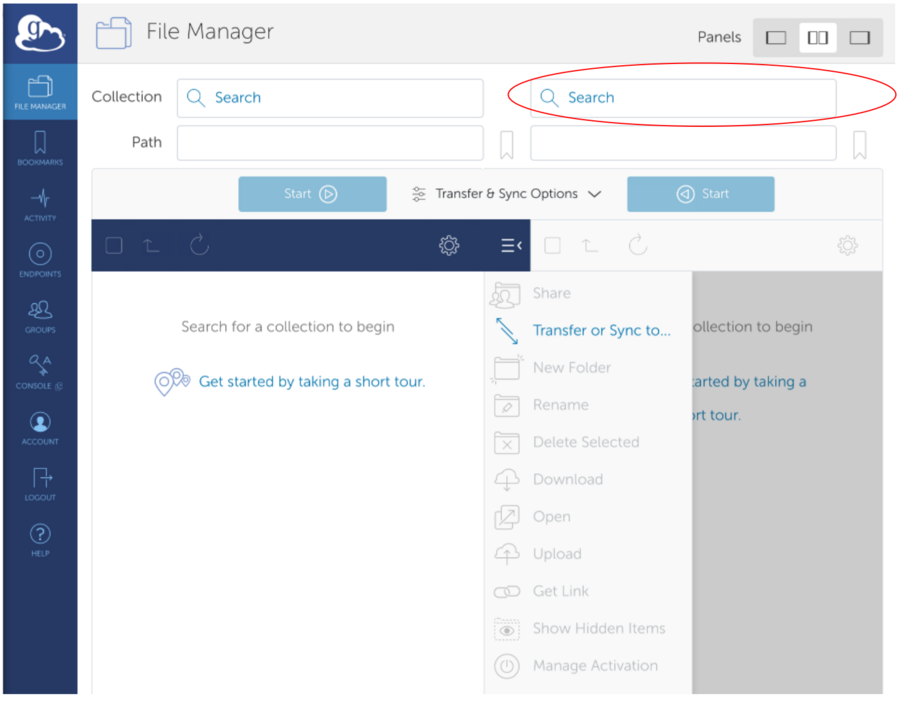
*Select Remote Endpoint provided over email - Go to the "Your Collections" or "Shared With You" tab
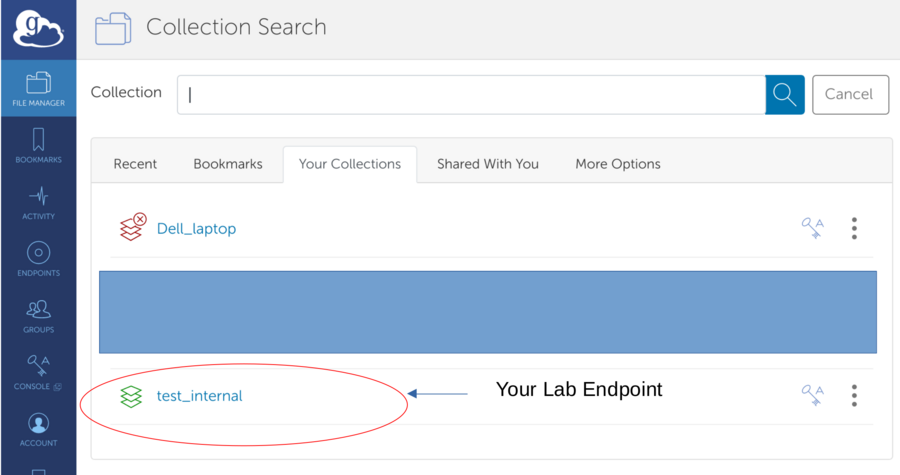
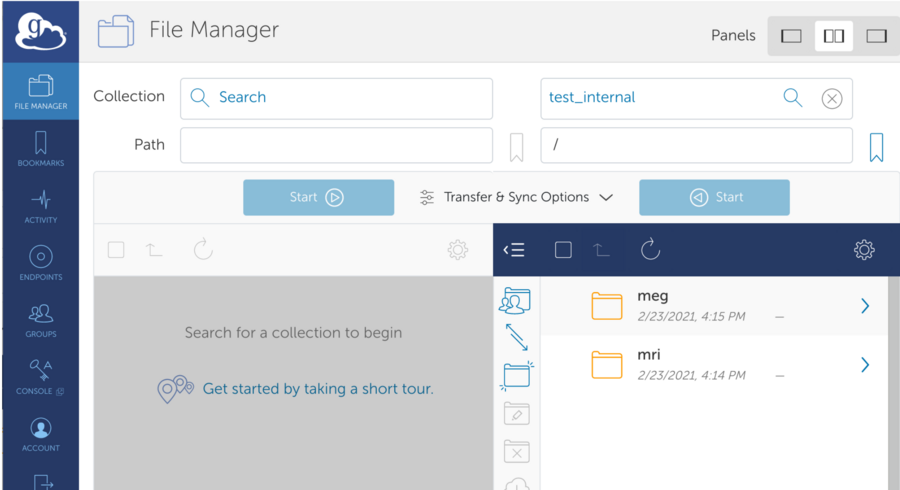
*Connected to Remote Endpoint - View Contents and select folder (It is possible that the folder is empty)
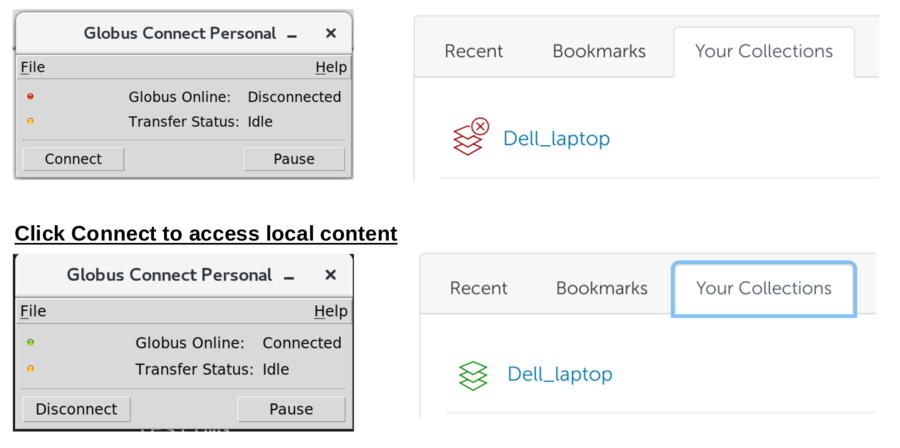
*Start Globus Connect Personal on your computer to interact with local files/folders.
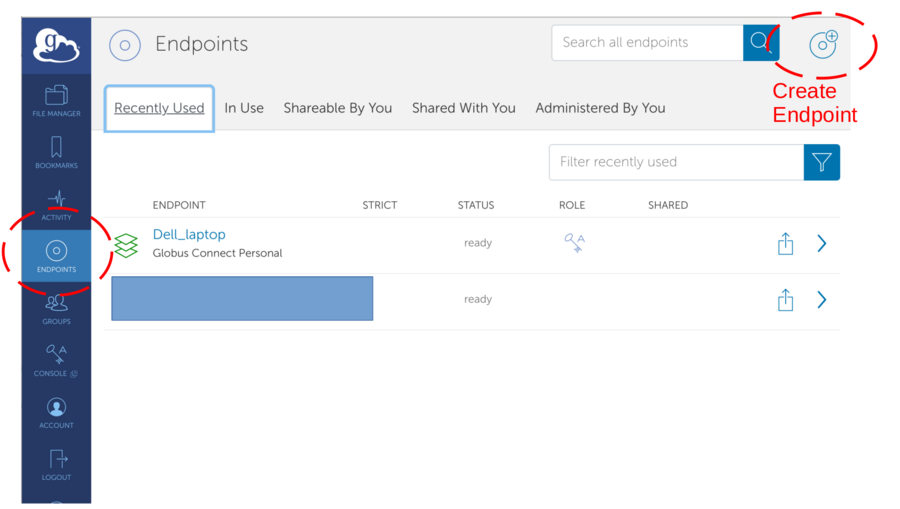
If no collection available - see next section to create endpoint
*Create a local endpoint (on your computer)
*Navigate Local Path. Select Data. Hit Start to initiate upload
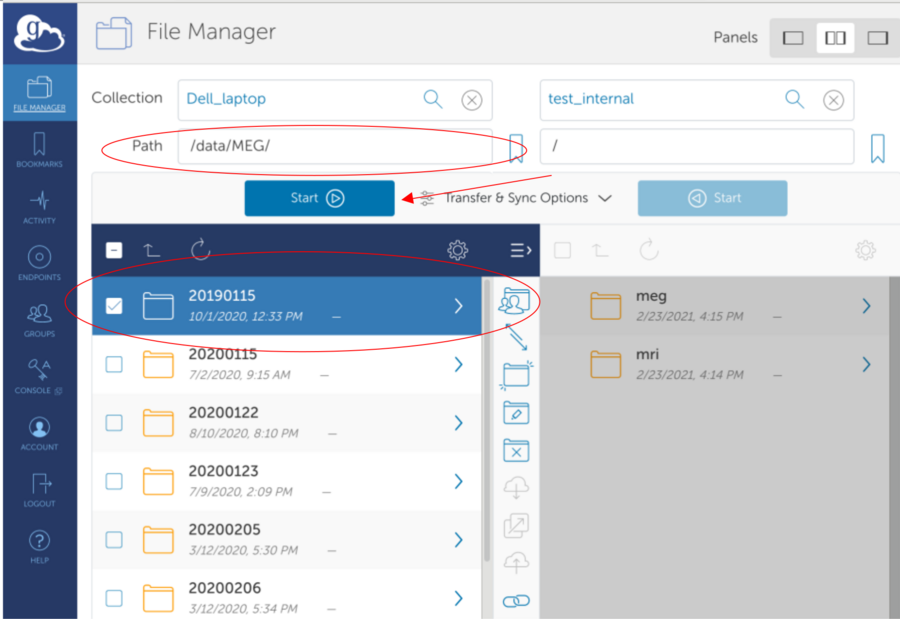
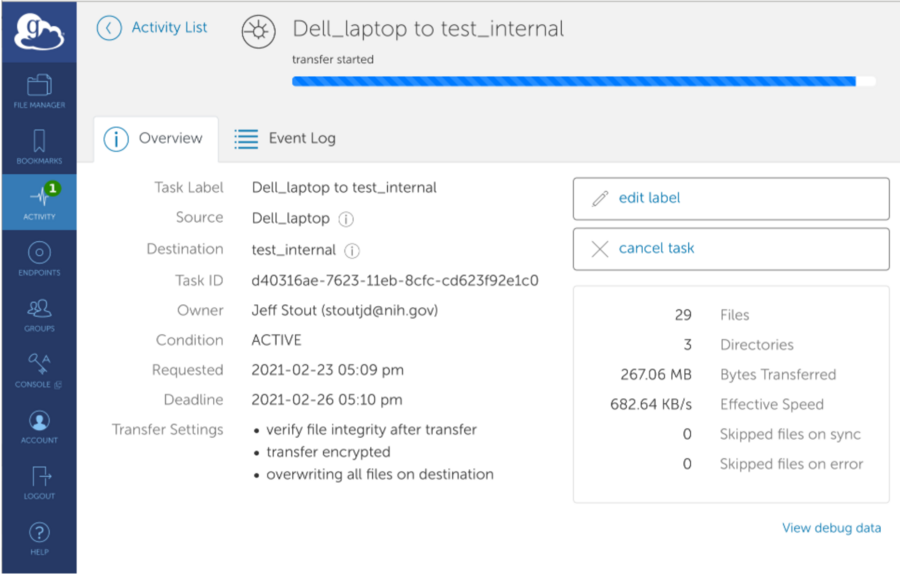
*After completion: Globus upload details and email of successful transfer