Mne bids pipeline: Difference between revisions
| (4 intermediate revisions by the same user not shown) | |||
| Line 60: | Line 60: | ||
sinteractive --mem=6G --cpus-per-task=4 --gres=lscratch:50 #adjust mem and cpus accordingly - subjects will run in parrallel |
sinteractive --mem=6G --cpus-per-task=4 --gres=lscratch:50 #adjust mem and cpus accordingly - subjects will run in parrallel |
||
===Create BIDS data from |
===Option 1: Create BIDS data from commandline=== |
||
module load mne_scripts |
module load mne_scripts |
||
| Line 71: | Line 71: | ||
make_meg_bids.py -meg_input_dir ${MEGFOLDER} -mri_bsight ${MRI_BSIGHT}.nii -mri_bsight_elec ${EXPORTED_BRAINSIGHT}.txt |
make_meg_bids.py -meg_input_dir ${MEGFOLDER} -mri_bsight ${MRI_BSIGHT}.nii -mri_bsight_elec ${EXPORTED_BRAINSIGHT}.txt |
||
===Option 2: Use CSV file to create BIDS (rest data only)=== |
|||
Instructions and more info: https://github.com/nih-megcore/enigma_anonymization_lite <br> |
|||
Fill out the csv with all of the appropriate entries <br> |
|||
module load enigma_meg |
|||
enigma_anonymization_mne.py -topdir TOPDIR -csvfile CSVFILE -njobs NJOBS -linefreq LINEFREQ -bidsonly #Use the bidsonly flag if not anonymizing |
|||
==MNE BIDS Pipeline== |
|||
===Freesurfer Processing=== |
===Freesurfer Processing=== |
||
If you have already processed your freesurfer data - just copy or link the freesurfer data(see below). You can also set the freesurfer directory in mne-bids-pipeline config file. Alternatively, you can have the pipieline calculate freesurfer as part of the processing. This will take time - adjust sinteractive session (see below). |
If you have already processed your freesurfer data - just copy or link the freesurfer data(see below). You can also set the freesurfer directory in mne-bids-pipeline config file. Alternatively, you can have the pipieline calculate freesurfer as part of the processing. This will take time - adjust sinteractive session (see below). |
||
| Line 92: | Line 99: | ||
#--steps=preprocessing,sensor,source,report or all (default) - Can be a list of steps or a single step |
#--steps=preprocessing,sensor,source,report or all (default) - Can be a list of steps or a single step |
||
#--subject=SUBJECTID(without the sub- prefix) |
#--subject=SUBJECTID(without the sub- prefix) |
||
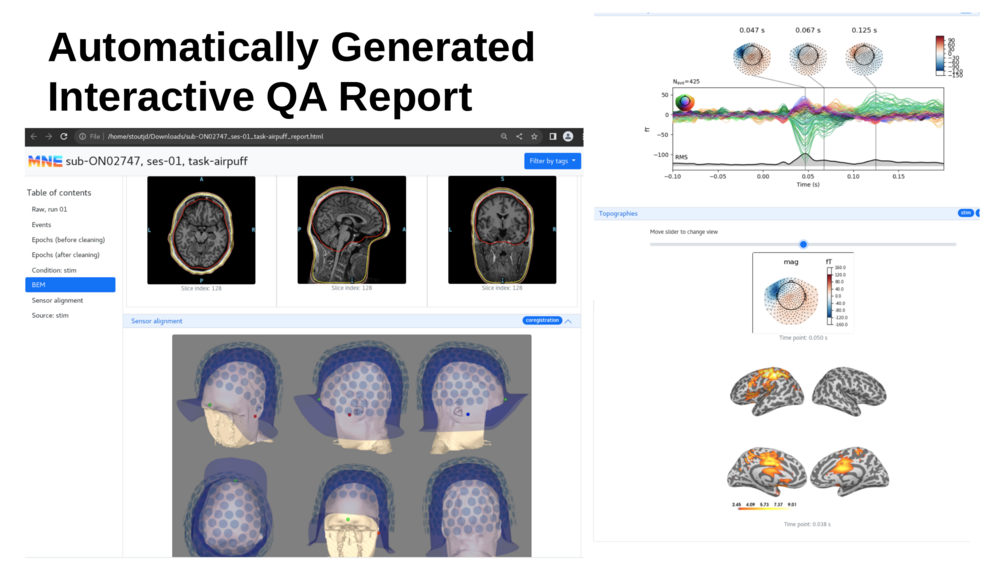
====Example Output Images (from anonymized data - the pipeline does not anonymize)==== |
|||
[[File : MNE BIDS PIPELINE output.png | 1000px ]] |
|||
==TEST DATA for biowulf== |
==TEST DATA for biowulf== |
||
| Line 115: | Line 125: | ||
''Open sub-ON02747_ses-01_task-airpuff_report.html in an internet browser to view the report'' |
''Open sub-ON02747_ses-01_task-airpuff_report.html in an internet browser to view the report'' |
||
==Setup for running on a headless server (no monitor attached)== |
|||
#(add this to the bashrc and run **start_xvfb** before running): |
|||
alias start_xvfb='Xvfb :99 & export DISPLAY=:99' |
|||
export MESA_GL_VERSION_OVERRIDE=3.3 |
|||
export MNE_3D_OPTION_ANTIALIAS=false |
|||
Latest revision as of 10:12, 3 July 2023
Skip Background
Intro
BIDS is a standard specification for neuroimaging/physiology data. This currently includes at least: MRI, fMRI, DTI, EEG, MEG, fNIRS (and possibly ECOG/sEEG). BIDS typically describes how RAW data is organized - and processed data is located in the bids_dir/derivatives/{AnalysisPackage}/{SUBJECT}/... The main advantage is that common code can be generated to process data organized in a standard format. Therefore, you should be able to import the bids data into any number of neurophysiological packages (MNE, Brainstorm, SPM, Fieldtrip, ...). Additionally, standardized processing packages known as BIDS apps can be used to process the data in the same way as long as the data is organized in BIDS.
BIDS (formal specs) >> MNE_BIDS (python neurophysiological BIDS) >> MNE_BIDS_PIPELINE (structured processing in MNE python according to BIDS definition)
BIDS website
https://bids-specification.readthedocs.io/en/stable/
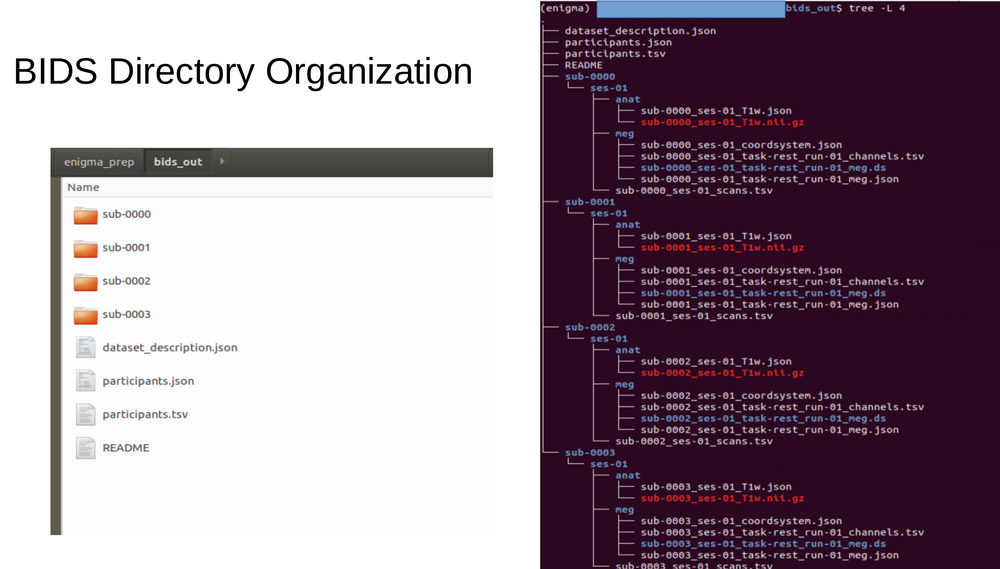
Bids data organization
MNE Bids
MNE bids is a python tool to make and access neurophysiological data. This tool is a branch of the MNE tools for neurophysiological analysis.
MNE Bids website
https://mne.tools/mne-bids/stable/index.html
MNE bids pipeline background
Mne bids pipeline website:
https://mne.tools/mne-bids-pipeline/
Full example:
https://mne.tools/mne-bids-pipeline/examples/ds000248.html
Processing
All processing is defined in the config.py file. This has hundreds of options to define the processing.
Here are some of the typical options: https://mne.tools/mne-bids-pipeline/settings/general.html
Here is the full list of options:
https://github.com/mne-tools/mne-bids-pipeline/blob/main/config.py
Example Config
(copy into a config.py text file and use)
Modify for use with your data - at a minimum: bids_root, conditions
study_name = 'TESTSTudy' bids_root = 'YOUR_BIDS_DIR' #<< Modify task = 'TASKNAME' #If this is left empty, it will likely find another task and throw an error l_freq = 1.0 h_freq = 100. epochs_tmin = -0.1 epochs_tmax = 0.2 baseline = (-0.1, 0.0) resample_sfreq = 300.0 ch_types = ['meg'] conditions = ['stim'] #<< list of conditions N_JOBS=4 #On biowulf - leave this empty - it will set N_JOBS equal to the cpu cores assigned in sinteractive on_error = 'continue' # This is often helpful when doing multiple subjects. If 1 subject fails processing stops #crop_runs = [0, 900] #Can be useful for long tasks that are ended early but full file is written.
Use on biowulf
!!If you run into any issues - please let Jeff Stout know so that the code can be updated!!
Make MEG modules accessible
#Run the line below every login
#Or add the following line to your ${HOME}/.bashrc (you will need to re-login or type bash to initialize)
module use --append /data/MEGmodules/modulefiles
Start interactive session with scratch to render visualization offscreen
sinteractive --mem=6G --cpus-per-task=4 --gres=lscratch:50 #adjust mem and cpus accordingly - subjects will run in parrallel
Option 1: Create BIDS data from commandline
module load mne_scripts
#Afni coregistered mri data
make_meg_bids.py -meg_input_dir ${MEGFOLDER} -mri_brik ${AFNI_COREGED}+orig.BRIK
## OR ##
#Brainsight coregistered mri data
make_meg_bids.py -meg_input_dir ${MEGFOLDER} -mri_bsight ${MRI_BSIGHT}.nii -mri_bsight_elec ${EXPORTED_BRAINSIGHT}.txt
Option 2: Use CSV file to create BIDS (rest data only)
Instructions and more info: https://github.com/nih-megcore/enigma_anonymization_lite
Fill out the csv with all of the appropriate entries
module load enigma_meg enigma_anonymization_mne.py -topdir TOPDIR -csvfile CSVFILE -njobs NJOBS -linefreq LINEFREQ -bidsonly #Use the bidsonly flag if not anonymizing
MNE BIDS Pipeline
Freesurfer Processing
If you have already processed your freesurfer data - just copy or link the freesurfer data(see below). You can also set the freesurfer directory in mne-bids-pipeline config file. Alternatively, you can have the pipieline calculate freesurfer as part of the processing. This will take time - adjust sinteractive session (see below).
Already Processed - Copy Data
mkdir -p ${bids_dir}/derivatives/freesurfer/subjects #If this folder doesn't already exists
#Copy your subject from the freesurfer directory to the bids DERIVATIVES freesurfer directory
cp -R ${SUBJECTS_DIR}/${SUBJID} ${bids_dir}/derivatives/freesurfer/subjects
Process Using MNE-BIDS-Pipeline
#your sinteractive session must include --time=24:00:00 so that the freesurfer processing doesn't time out #add the following step to your mne_bids_pipeline processing --steps=freesurfer
Process data using MNE Bids Pipeline
First make a config file: example
module purge
module load mne_bids_pipeline
mne-bids-pipeline-run.py --config=CONFIG.py
#Optional Flags
#--steps=preprocessing,sensor,source,report or all (default) - Can be a list of steps or a single step
#--subject=SUBJECTID(without the sub- prefix)
Example Output Images (from anonymized data - the pipeline does not anonymize)
TEST DATA for biowulf
This section provides some test data to analyze using mne-bids-pipeline. Feel free to adjust parameters in the config.py after you run through the analysis the first time. All of the analysis parameters are defined in the config.py provided. Additional config.py parameters can be found above in Processing.
Start Interactive Session
sinteractive --mem=6G --cpus-per-task=4 --gres=lscratch:50
Copy and untar the data into your folder
cp -R /vf/users/MEGmodules/modules/bids_example_data_airpuff.tar ./ tar -xvf bids_example_data_airpuff.tar #Add the bids_root to your config file echo bids_root=\'$(pwd)/bids_example_data_airpuff\' >> $(pwd)/bids_example_data_airpuff/config.py
Load module and process the data
module load mne_bids_pipeline mne-bids-pipeline-run.py --config=$(pwd)/bids_example_data_airpuff/config.py
#Copy this path for the next step echo $(pwd)/bids_example_data_airpuff/derivatives/mne-bids-pipeline/sub-ON02747/ses-01/meg/sub-ON02747_ses-01_task-airpuff_report.html
#In another terminal download your results from biowulf
scp ${USERNAME}@helix.nih.gov:${PathFromAbove} ./
Open sub-ON02747_ses-01_task-airpuff_report.html in an internet browser to view the report
Setup for running on a headless server (no monitor attached)
#(add this to the bashrc and run **start_xvfb** before running): alias start_xvfb='Xvfb :99 & export DISPLAY=:99' export MESA_GL_VERSION_OVERRIDE=3.3 export MNE_3D_OPTION_ANTIALIAS=false