ENIGMA MEG Working Group: Difference between revisions
mNo edit summary |
mNo edit summary |
||
| (6 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
UNDER CONSTRUCTION |
|||
| ⚫ | |||
[http://enigma.ini.usc.edu |
[http://enigma.ini.usc.edu/ The ENIGMA Consortium]<br> |
||
| ⚫ | |||
==Enigma Project - MEG working group== |
==Enigma Project - MEG working group== |
||
The goal of the ENIGMA MEG Working Group is to explore the spatiotemporal patterns of brain oscillatory activity, and determine how these patterns relate to age, |
The goal of the ENIGMA MEG Working Group is to explore the spatiotemporal patterns of brain oscillatory activity, and determine how these patterns relate to age, sex, and common genetic variants. Data from numerous modalities have demonstrated a hierarchical organization of the brain from sensory systems to higher cortical areas. There is some evidence that this hierarchical organization may also be reflected in the spatial distribution of intrinsic timescales of activity. This ENIGMA working group intends to explore these patterns across the lifespan. To achieve the highest possible number of subjects, this needs to be done by meta-analysis.<br> |
||
[mailto:nugenta@nih.gov Allison Nugent, PhD], ENIGMA MEG Chair <br> |
[mailto:nugenta@nih.gov Allison Nugent, PhD], ENIGMA MEG Chair <br> |
||
[mailto:jeff.stout@nih.gov Jeff Stout, PhD], Lead Scientist <br> |
[mailto:jeff.stout@nih.gov Jeff Stout, PhD], Lead Scientist <br> |
||
[mailto:anna.namyst@nih.gov Anna Namyst], Project Coordinator</br> |
[mailto:anna.namyst@nih.gov Anna Namyst, BA], Project Coordinator</br> |
||
==MEG Working Group: Participating Institutions== |
==MEG Working Group: Participating Institutions== |
||
[[File:ENIGMA-MEG-WG 12.2021.jpg|frameless|right|500px|600px| Global Participation in the ENIGMA MEG Working Group]] |
|||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
* Aston University, UK |
|||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
* Advent Health for Children |
* Advent Health for Children |
||
| ⚫ | |||
| ⚫ | |||
* University |
* Aston University, UK |
||
| ⚫ | |||
| ⚫ | |||
* Cambridge University |
* Cambridge University |
||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
* DNISC (Università degli Studi G.D'Annunzio Chieti Pescara) |
* DNISC (Università degli Studi G.D'Annunzio Chieti Pescara) |
||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
* Johannes Gutenberg University, Mainz, Germany (Biomedical statistics and multimodal signal processing Unit) |
* Johannes Gutenberg University, Mainz, Germany (Biomedical statistics and multimodal signal processing Unit) |
||
| ⚫ | |||
* Macquarie University |
* Macquarie University |
||
| ⚫ | |||
| ⚫ | |||
* Massachusetts General Hospital |
* Massachusetts General Hospital |
||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
* Precision Medicine Centre, Hokuto Hospital |
* Precision Medicine Centre, Hokuto Hospital |
||
* Precision Medicine Centre, Kumagaya General Hospital |
* Precision Medicine Centre, Kumagaya General Hospital |
||
| ⚫ | |||
| ⚫ | |||
* Thomas Jefferson University |
|||
* Universiti Sains Malaysia |
* Universiti Sains Malaysia |
||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
* University of Texas Southwestern |
|||
* University of Tübingen |
|||
If you are interested in participating in this project, please email the project coordinator, [mailto:anna.namyst@nih.gov Anna Namyst], for more information. |
If you are interested in participating in this project, please email the project coordinator, [mailto:anna.namyst@nih.gov Anna Namyst], for more information. |
||
==Data Analysis== |
==Data Analysis== |
||
Data analysis will be available in two flavors: |
Data analysis will be available in two flavors: |
||
1. Upload coregistered anonymized data to NIH |
|||
2. Perform analysis at acquisition site using EnigmaMeg scripts |
|||
*[https://github.com/jstout211/enigma_MEG ENIGMA MEG Pipeline on GitHub] |
|||
*[https://megcore.nih.gov/index.php/ENIGMA_MEG_Pipeline_FAQ Processing Pipeline FAQ] |
|||
==Upload Data to be analyzed on NIH Biowulf cluster== |
|||
Instructions for data |
Instructions for data preparation and upload: <br> |
||
[ |
[https://megcore.nih.gov/index.php/ENIGMA_MEG_data_upload ENIGMA MEG Data Upload] |
||
==Potential MEG Subanalyses== |
|||
Healthy Volunteers |
Healthy Volunteers |
||
Epilepsy |
Epilepsy |
||
Latest revision as of 15:39, 31 January 2025
The ENIGMA Consortium
The ENIGMA MEG Working Group
Enigma Project - MEG working group
The goal of the ENIGMA MEG Working Group is to explore the spatiotemporal patterns of brain oscillatory activity, and determine how these patterns relate to age, sex, and common genetic variants. Data from numerous modalities have demonstrated a hierarchical organization of the brain from sensory systems to higher cortical areas. There is some evidence that this hierarchical organization may also be reflected in the spatial distribution of intrinsic timescales of activity. This ENIGMA working group intends to explore these patterns across the lifespan. To achieve the highest possible number of subjects, this needs to be done by meta-analysis.
Allison Nugent, PhD, ENIGMA MEG Chair
Jeff Stout, PhD, Lead Scientist
Anna Namyst, BA, Project Coordinator
MEG Working Group: Participating Institutions
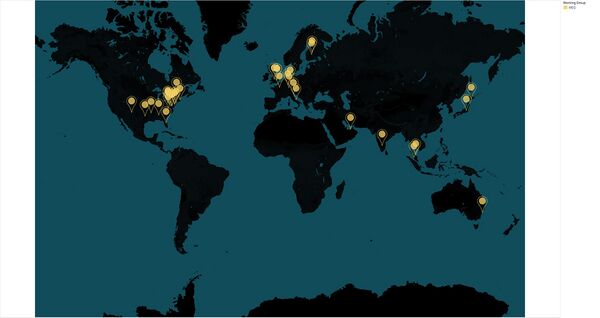
- Advent Health for Children
- Arkansas Childrens Hospital
- Aston University, UK
- Cambridge University
- Children's Hospital of Philadephia
- DNISC (Università degli Studi G.D'Annunzio Chieti Pescara)
- IRCCS San Camillo Hospital Venice
- Johannes Gutenberg University, Mainz, Germany (Biomedical statistics and multimodal signal processing Unit)
- Macquarie University
- Massachusetts General Hospital
- McGill University, Montreal Neurological Institute
- National Institute of Mental Health and Neurosciences (NIMHANS), Bengaluru, Karnataka, India
- Nemours DuPont Hospital for Children
- NeuroSpin
- New York University and NYU Abu Dhabi
- NIH/NICHD
- NIH/NIMH
- Precision Medicine Centre, Hokuto Hospital
- Precision Medicine Centre, Kumagaya General Hospital
- Salisbury VA Medical Center, Wake Forest University, Wake Forest School of Medecine
- The Mind Research Network
- Thomas Jefferson University
- Universiti Sains Malaysia
- Universiti Teknologi PETRONAS
- University of Alabama, Birmingham
- University of Göttingen
- University of Jyväskylä, Finland
- University of Pittsburgh
- University of Pittsburgh
- University of Texas Southwestern
- University of Tübingen
If you are interested in participating in this project, please email the project coordinator, Anna Namyst, for more information.
Data Analysis
Data analysis will be available in two flavors: 1. Upload coregistered anonymized data to NIH 2. Perform analysis at acquisition site using EnigmaMeg scripts
Upload Data to be analyzed on NIH Biowulf cluster
Instructions for data preparation and upload:
ENIGMA MEG Data Upload
Potential MEG Subanalyses
Healthy Volunteers Epilepsy Alzheimer's and dementia Motor Analysis Language Processing Anxiety Disorders Schizophrenia and related disorders Developmental disorders Traumatic Brain Injury Stroke