Meg analysis pipeline: Difference between revisions
No edit summary |
|||
| (6 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
!!! UNDER CONSTRUCTION !!! |
|||
====Reproducible MEG Analysis==== |
====Reproducible MEG Analysis==== |
||
MEG data collection and analysis requires a lot of interacting components (Stimulus delivery, logfiles, projector onset triggers, analog and digital triggering, electrophysiology measures, MRI coregistration...) that can lead to heterogeneous data and data quality. This makes pipeline analysis particularly difficult due to the variability in inputs. Additionally, with the multitude of techniques and metrics to evaluate MEG data (dipole/MNE/dSPM/beamformer/DICS beamformer/parametric stats/non-parametric stats ....) projects can have evolving analysis protocols. Knowing what preprocessing was performed on what datasets can often come down to an excel spreadsheet or text document. Since these projects can sometimes last multiple years, it is important to know what was done to produce a specific result and how to apply these analysis techniques to new incoming data. |
MEG data collection and analysis requires a lot of interacting components (Stimulus delivery, logfiles, projector onset triggers, analog and digital triggering, electrophysiology measures, MRI coregistration...) that can lead to heterogeneous data and data quality. This makes pipeline analysis particularly difficult due to the variability in inputs. Additionally, with the multitude of techniques and metrics to evaluate MEG data (dipole/MNE/dSPM/beamformer/DICS beamformer/parametric stats/non-parametric stats ....) projects can have evolving analysis protocols. Knowing what preprocessing was performed on what datasets can often come down to an excel spreadsheet or text document. Since these projects can sometimes last multiple years, it is important to know what was done to produce a specific result and how to apply these analysis techniques to new incoming data. |
||
===Check Inputs=== |
|||
#Verify that inputs are present |
|||
#Find raw MRI, raw MEG, logfiles |
|||
#Test example below - this is automated if MRIpath/MEGpath/LogfilePath set |
|||
num_mri = len(glob.glob(os.path.join(mri_path, subjid)) |
|||
print('{} has {} MRIs present'.format(subjid, num_mri)) |
|||
===QA datasets=== |
|||
#Create QA scripts for each task |
|||
assert len(2back) == 40 |
|||
assert len(correct_response) > 20 |
|||
===Containerized Computing Environment=== |
===Containerized Computing Environment=== |
||
| Line 27: | Line 43: | ||
===Nipype=== |
===Nipype=== |
||
[https://nipype.readthedocs.io/en/latest/ Nipype Pipeline Toolbox] |
[https://nipype.readthedocs.io/en/latest/ Nipype Pipeline Toolbox] </br></br> |
||
Nipype uses compute modules that can stitch together Bash, Python, and Matlab calls. There are also modules available for all major MRI/fMRI software packages (afni, spm, freesurfer, fsl ...). These modules produce a defined input/output requirements. For example a pipeline will fail all dependency processes if a required output file is not generated. We have developed nipype modules for the major computing components of our data analysis pipeline and have some general analysis pipelines available for the NIMH data (see image below). Nipype additionally documents all output provenance at runtime - including shell variables and computing environment. </br> |
|||
| ⚫ | |||
[[File:Nipype pipeline.png| center | 400px]] </br> |
|||
| ⚫ | |||
===Git=== |
===Git=== |
||
Latest revision as of 12:04, 21 August 2020
!!! UNDER CONSTRUCTION !!!
Reproducible MEG Analysis
MEG data collection and analysis requires a lot of interacting components (Stimulus delivery, logfiles, projector onset triggers, analog and digital triggering, electrophysiology measures, MRI coregistration...) that can lead to heterogeneous data and data quality. This makes pipeline analysis particularly difficult due to the variability in inputs. Additionally, with the multitude of techniques and metrics to evaluate MEG data (dipole/MNE/dSPM/beamformer/DICS beamformer/parametric stats/non-parametric stats ....) projects can have evolving analysis protocols. Knowing what preprocessing was performed on what datasets can often come down to an excel spreadsheet or text document. Since these projects can sometimes last multiple years, it is important to know what was done to produce a specific result and how to apply these analysis techniques to new incoming data.
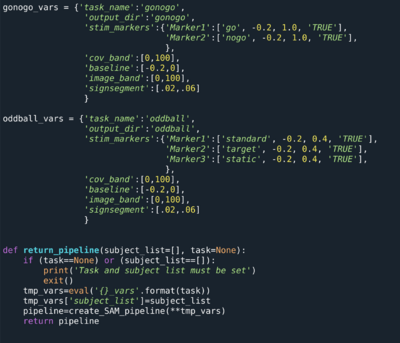
Check Inputs
#Verify that inputs are present
#Find raw MRI, raw MEG, logfiles
#Test example below - this is automated if MRIpath/MEGpath/LogfilePath set
num_mri = len(glob.glob(os.path.join(mri_path, subjid))
print('{} has {} MRIs present'.format(subjid, num_mri))
QA datasets
#Create QA scripts for each task assert len(2back) == 40 assert len(correct_response) > 20
Containerized Computing Environment
#It can be easy to build a container from a current conda environment file conda env export >> environment.yml #It may be necessary to remove some components such as VTK
#Create a singularity def file (textfile) from below
Bootstrap: docker
From: continuumio/miniconda3
%files
environment.yml
%post
/opt/conda/bin/conda env update --file environment.yml --name base
%runscript
exec "$@"
#Build container using singularity sudo singularity build conda_env.sif def_file.def
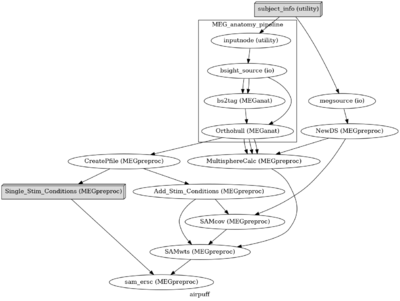
Nipype
Nipype Pipeline Toolbox
Nipype uses compute modules that can stitch together Bash, Python, and Matlab calls. There are also modules available for all major MRI/fMRI software packages (afni, spm, freesurfer, fsl ...). These modules produce a defined input/output requirements. For example a pipeline will fail all dependency processes if a required output file is not generated. We have developed nipype modules for the major computing components of our data analysis pipeline and have some general analysis pipelines available for the NIMH data (see image below). Nipype additionally documents all output provenance at runtime - including shell variables and computing environment.