Analysis with Nipype: Difference between revisions
Jump to navigation
Jump to search
Content added Content deleted
(Created page with "'''Nipype is a pipeline configuration tool that controls analysis flow and easily reconfigures for added functionality'''<br> Nipype has strict requirements on inputs and out...") |
|||
| (13 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
'''UNDER CONSTRUCTION'''<br> |
|||
'''Nipype is a pipeline configuration tool that controls analysis flow and easily reconfigures for added functionality |
'''Nipype is a pipeline configuration tool that controls analysis flow and easily reconfigures for added functionality. |
||
Nipye controls the flow of data and automatically determines dependencies to create filenames and folders'''<br> |
|||
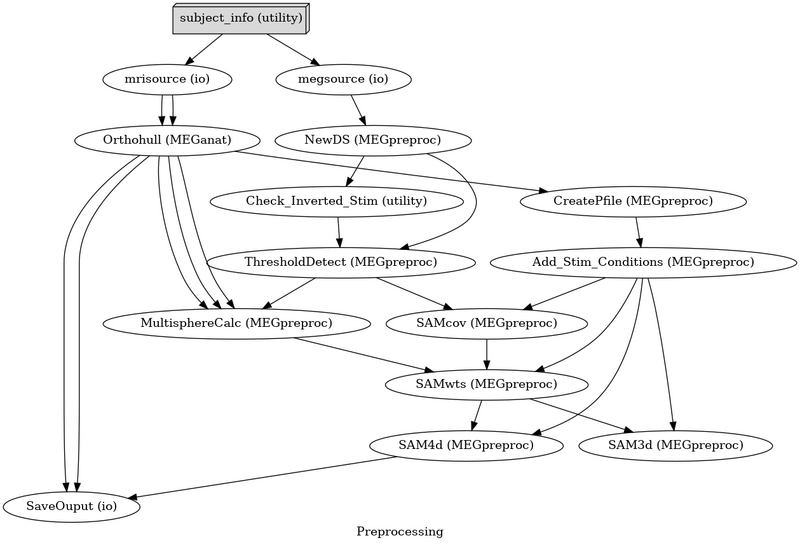
[[Image:Nipype meg.png | 800px]] |
|||
===Nipype Resources=== |
|||
https://nipype.readthedocs.io/en/latest/ <br> |
|||
https://miykael.github.io/nipype_tutorial/ <br> |
|||
===Nipype configuration for Biowulf=== |
|||
pull pyctf <br> |
|||
module load fftw/3.3.8/openmpi-4.0.0/gcc-7.3.0 ## it is possible to use another version - do 'module spider fftw' |
|||
cd $PYCTF_DIR #Folde will have the Makefile |
|||
make clean |
|||
make |
|||
make install |
|||
make usersite (will add a link so package is seen in conda) |
|||
pull SAMsrcV5 <br> |
|||
For installation instructions read the INSTALL text file - somewhat summarized below |
|||
cd $SAMSRCV5_DIR |
|||
make |
|||
cd config |
|||
make symlinks |
|||
pull megblocks <br> |
|||
install miniconda (or Anaconda) <br> |
|||
https://hpc.nih.gov/apps/python.html#envs << Go to the Creating Python Environments Section to configure |
|||
*Add environment information to miniconda ##Need to provide environment.yml |
|||
*Add pyctf, SAMsrc, and megblocks to the miniconda environment ## TODO < provide a way to do this. Add the paths to miniconda/env......pth file |
|||
===Running Pipeline=== |
|||
Login to biowulf |
|||
# --cpus-per-task=Multithreading amount |
|||
# --ntasks=Number of Parrallel tasks |
|||
# Total CPU count = (cpus-per-task * ntasks) - Most nodes support 56 max CPUs |
|||
sinteractive --exclusive --cpus-per-task=8 --mem-per-cpu=2g --ntasks=6 |
|||
export OMP_NUM_THREADS=4 #Otherwise these tasks will try to use all of the cores |
|||
module load afni |
|||
module load ctf |
|||
conda activate mne #This is whatever environment configured for nipype |
|||
~/ |
|||
Nipype has strict requirements on inputs and outputs <br> |
Nipype has strict requirements on inputs and outputs <br> |
||
| Line 13: | Line 57: | ||
# Resting State |
# Resting State |
||
## |
## |
||
SAM input base class used for all of the SAM functions |
|||
Configuration on Biowulf: |
|||
# Set up conda: instructions on NIH HPC - https://hpc.nih.gov/apps/python.html |
|||
# Activate the conda environment and install nipype |
|||
#If you do not know your environment name type: conda env list |
|||
conda activate $ENVIRONMENT_NAME |
|||
conda install -c nipype conda-forge |
|||
Latest revision as of 11:22, 7 February 2020
UNDER CONSTRUCTION
Nipype is a pipeline configuration tool that controls analysis flow and easily reconfigures for added functionality.
Nipye controls the flow of data and automatically determines dependencies to create filenames and folders
Nipype Resources
https://nipype.readthedocs.io/en/latest/
https://miykael.github.io/nipype_tutorial/
Nipype configuration for Biowulf
pull pyctf
module load fftw/3.3.8/openmpi-4.0.0/gcc-7.3.0 ## it is possible to use another version - do 'module spider fftw' cd $PYCTF_DIR #Folde will have the Makefile make clean make make install make usersite (will add a link so package is seen in conda)
pull SAMsrcV5
For installation instructions read the INSTALL text file - somewhat summarized below cd $SAMSRCV5_DIR make cd config make symlinks
pull megblocks
install miniconda (or Anaconda)
https://hpc.nih.gov/apps/python.html#envs << Go to the Creating Python Environments Section to configure *Add environment information to miniconda ##Need to provide environment.yml
- Add pyctf, SAMsrc, and megblocks to the miniconda environment ## TODO < provide a way to do this. Add the paths to miniconda/env......pth file
Running Pipeline
Login to biowulf
# --cpus-per-task=Multithreading amount # --ntasks=Number of Parrallel tasks # Total CPU count = (cpus-per-task * ntasks) - Most nodes support 56 max CPUs sinteractive --exclusive --cpus-per-task=8 --mem-per-cpu=2g --ntasks=6 export OMP_NUM_THREADS=4 #Otherwise these tasks will try to use all of the cores module load afni module load ctf conda activate mne #This is whatever environment configured for nipype ~/
Nipype has strict requirements on inputs and outputs
- InputSpec
- OutputSpec
Preconfigured pipeline templates:
- Epilepsy
- Sam_epi
- Task
- Parsemarks, addmarks, samcov, sam3d, ... stats?
- Resting State
SAM input base class used for all of the SAM functions
Configuration on Biowulf:
- Set up conda: instructions on NIH HPC - https://hpc.nih.gov/apps/python.html
- Activate the conda environment and install nipype
#If you do not know your environment name type: conda env list conda activate $ENVIRONMENT_NAME conda install -c nipype conda-forge