Brainsight Coregistration: Difference between revisions
No edit summary |
|||
| (41 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
== OVERVIEW == |
|||
BRAINSIGHT COREGISTRATION QUICK-START GUIDE [UNDER CONSTRUCTION] |
|||
Brainsight is a useful tool for modeling anatomical data for coregistration with the MEG data collected during your study. With Brainsight, you are able to set anatomical landmarks, generate a 3D rending of the head, and mark the locations of MEG marker coils and EEG electrodes. Coregistration can be done quickly before your recording and eliminates the need to do it by hand in AFNI at a later date (although, we recommend verifying your coregistration in AFNI or other similar software as you go along). |
|||
This guide is intended to be a resource for MEG Core users who have already been trained to use Brainsight in our lab. If you would like to schedule training, you can reach out to Anna Namyst directly or submit a request via [https://kurage.nimh.nih.gov/nih/staff/form.html the Staff Support Request form.] |
|||
OVERVIEW |
|||
[[File:Brainsight Coregistration Flowchart|frame|center|Overview of Brainsight Coregistration ]] |
|||
To learn more about Brainsight, you can read more [https://www.rogue-research.com/tms/brainsight-tms/ here]. |
|||
[[File:BrainsighCoregistration-Flowchart.jpg|frame|center|upright=1.0|Overview of Brainsight coregistration for standard MEG studies]] |
|||
== PREPARATION == |
== PREPARATION == |
||
| Line 11: | Line 13: | ||
=== Downloading/Preparing the MRI data === |
=== Downloading/Preparing the MRI data === |
||
If your participant’s data was collected here prior to October 2022, you can download their data from the FMRIF DICOM Archive: |
|||
https://gold.nimh.nih.gov/ |
|||
If your participant’s data was collected here '''AFTER October 2022''', you can download their data from the FMRIF XNAT Archive: |
|||
* Transform the DICOM to NIfTI (.nii) format: |
|||
https://fmrif-xnat.nimh.nih.gov/ |
|||
* Locate the MP RAGE ''(The README file from the session will tell you which directory to use.)'' |
|||
If you don’t have access to either data repository – talk to your PI. |
|||
* WITHIN that directory-- |
|||
* Command: Dimon -infile_prefix anat_t1w_mp_rage_1mm_abcd -GERT_Rico -gert_write_as_nifti –gert_create_dataset |
|||
==== Transform the DICOM to NIfTI (.nii) format ==== |
|||
If your subject doesn’t have an MPRAGE, you can replace the prefix ‘anat_t1w_mp_rage_1mm_abcd’ with the prefix of a suitable alternative (i.e. SPGR). If you’re unsure, consult your PI or MEG Core staff.) |
|||
* Locate the MP RAGE |
|||
The README file from the session will tell you which directory to use. |
|||
If your subject doesn’t have an MPRAGE, you can use a suitable alternative (i.e. SPGR). |
|||
If you’re unsure, consult your PI or MEG Core staff. |
|||
* Navigate to that directory in terminal and use the following command: |
|||
Dimon -infile_prefix anat_t1w_mp_rage_1mm_abcd -GERT_Rico -gert_write_as_nifti –gert_create_dataset |
|||
(If you use something other than MPRAGE, replace the prefix ‘anat_t1w_mp_rage_1mm_abcd’ accordingly.) |
|||
For more information on Dimon-- https://afni.nimh.nih.gov/pub/dist/doc/program_help/Dimon.html |
|||
For more information on Dimon-- https://afni.nimh.nih.gov/pub/dist/doc/program_help/Dimon.html |
|||
Generate MEG Hashcode for your subject |
|||
* Generate MEG Hashcode for your subject |
|||
Note: If you don’t have the opportunity to do it before your study, label your file something generic (I.e. SubjectXDate.nii). You will be able to rename it later; the primary goal is to remove the PII that NMRIF includes in the filename. PII may still be buried in the header text of the data; treat the files accordingly. |
|||
If you don’t have the opportunity to do it before your study, label your file something generic (I.e. SubjectXDate.nii). |
|||
You will be able to rename it later; the primary goal is to remove the PII that NMRIF includes in the filename. |
|||
PII may still be buried in the header text of the data; treat the files accordingly. |
|||
=== Transferring data to the Brainsight Mac === |
=== Transferring data to the Brainsight Mac === |
||
Transfer your data to your directory on tako |
* Transfer your data to your directory on tako |
||
* Transfer your data from tako to one of Linux computers in the MEG Lab (namako or kani) |
|||
scp -r [SOURCE]@tako.nimh.nih.gov:/tmp/[MRI data] [DESTINATION] |
|||
Transfer your data from tako to one of Linux computers in the MEG Lab (namako or kani) |
|||
* Using the dedicated MEG Lab flash drive, transfer your data to the Mac and file it in the appropriate MEGLAB directory for your study |
|||
scp -r [SOURCE]@tako.nimh.nih.gov:/tmp/[MRI data] [DESTINATION] |
|||
Using the dedicated MEG Lab flash drive, transfer your data to the Mac and file it in the appropriate MEGLAB directory for your study |
|||
== BRAINSIGHT COREGISTRATION == |
== BRAINSIGHT COREGISTRATION == |
||
| Line 41: | Line 52: | ||
=== Importing data (May be done before Subject arrives) === |
=== Importing data (May be done before Subject arrives) === |
||
Launch the Brainsight program and select “New Empty Project”. |
* Launch the Brainsight program and select “New Empty Project”. Note: If you do not have access to your subject’s MRI, choose “New MNI Head Project” and follow the Brainsight User Manual instructions for using an average brain template (see chapter 6). This is generally not recommended for our analyses |
||
Note: If you do not have access to your subject’s MRI, choose “New MNI Head Project” and follow the Brainsight User Manual instructions for using an average brain template (see chapter 6). This is generally not recommended for our analyses. |
|||
Select subject’s anatomical data |
|||
NIfTI files are recommended by MEG Core, but other accepted formats are MINC, Analyze, DICOM, PAR/REC, or BrainVoyager VMR. |
* Select subject’s anatomical data. NIfTI files are recommended by MEG Core, but other accepted formats are MINC, Analyze, DICOM, PAR/REC, or BrainVoyager VMR. |
||
When the data is loaded, select “Show Image and Details” to view the metadata and image volume, or proceed to the next step. |
*When the data is loaded, select “Show Image and Details” to view the metadata and image volume, or proceed to the next step. |
||
=== Automatic Skin Reconstruction === |
=== Automatic Skin Reconstruction === |
||
From the 3D Reconstruction tab, select “Skin” from the “New” drop-down menu. |
* From the 3D Reconstruction tab, select “Skin” from the “New” drop-down menu. |
||
* |
|||
If needed, adjust threshold and select “keep largest piece” to isolate the head from any MR noise |
* If needed, adjust threshold and select “keep largest piece” to isolate the head from any MR noise |
||
=== Setting anatomical landmarks === |
=== Setting anatomical landmarks === |
||
Note: These are anatomical landmarks clearly identifiable from the MRI. You are not placing the marker coils at this step. |
''Note: These are anatomical landmarks clearly identifiable from the MRI. You are not placing the marker coils at this step.'' |
||
From the “Landmarks” tab, select “Configure Landmarks”. |
* From the “Landmarks” tab, select “Configure Landmarks”. The landmarks window will display the Skin reconstruction and coronal, sagittal, and transverse slices. The cursor cross-hairs will be displayed in all four image windows. |
||
* With the cursor cross-hairs identify the Nasion, LPA (left preauricular), and RPA (right preauricular). Rather than using the tragus, we will use the supratrageal notch -- notch above the tragus where the tragus joins the upper ear. This point is less likely to be distorted in the MRI by earplugs or head pads. |
|||
The landmarks window will display the Skin reconstruction and coronal, sagittal, and transverse slices. The cursor cross-hairs will be displayed in all four image windows. |
|||
* For additional precision, you can select a fourth point on the tip of the nose or the outer canthus. You may use whatever point as long as it is unambiguously identifiable and fixed. |
|||
With the cursor cross-hairs identify the Nasion, LPA (left preauricular), and RPA (right preauricular). Rather than using the tragus, we will use the notch above the tragus where the tragus joins the upper ear. |
|||
* Now you are ready to start a new session. At this point, you can suspend your project or move directly to the registration session. |
|||
This point is less likely to be distorted in the MRI by earplugs or head pads. |
|||
* If you will be using OPMs, proceed to the Targets tab and complete the target trajectory setup for the OPM rig before moving to the registration session: |
|||
For additional precision, you can select a fourth point on the tip of the nose or the outer canthus. You may use whatever point as long as it is unambiguously identifiable and fixed. |
|||
** [[Brainsight Coregistration: OPM Rig Setup|Instructions for OPM Rig Setup]] |
|||
Now you are ready to start a new session. At this point, you can suspend your project or move directly to the registration session. |
|||
=== Subject Preparation === |
=== Subject Preparation === |
||
''Personal protective equipment '''must''' be worn from this point onward.'' The researcher (you) must wear gloves, and the participant must put on a skull cap before wearing the tracker headband. After the session, swab the stylus and the tracker band with alcohol. |
|||
Draw the marker coil locations on the subject. |
|||
*Draw the marker coil locations on the subject. |
|||
1.5cm above the nasion, 1.5cm from the tragus on a line with the outer canthus of the eye. |
|||
*1.5cm above the nasion, 1.5cm from the tragus on a line with the outer canthus of the eye. |
|||
Give the participant a skull cap and place the subject tracker headband on their head. |
|||
For EEG subjects who have already been gelled, use a green surgical cap. |
*Give the participant a skull cap and place the subject tracker headband on their head. For EEG subjects who have already been gelled, use a green surgical cap, but try to get it as close to the scalp as possible. |
||
Position them in the chair and select “New Online Session” from the session window. To select an existing session, highlight the desired session and select “Resume Session”. |
*Position them in the chair and select “New Online Session” from the session window. To select an existing session, highlight the desired session and select “Resume Session”. |
||
=== Registration Session === |
=== Registration Session === |
||
| Line 88: | Line 94: | ||
With the Session window open, go through the following tabs(steps): |
With the Session window open, go through the following tabs(steps): |
||
Targets (optional) |
==== Targets (optional) ==== |
||
The anatomical landmarks selected can be imported here, but this section was intended for other applications (i.e. TMS, surgery). |
The anatomical landmarks selected can be imported here, but this section was intended for other applications (i.e. TMS, surgery). |
||
This section may be updated as we find applications for this section. |
|||
Polaris |
==== Polaris ==== |
||
Ensure that the Polaris Vicra is on and operational. We recommend turning on the Polaris as soon as you arrive so it has time to warm up. |
|||
Troubleshooting: |
|||
Ensure that the Polaris Vicra is on and operational. |
|||
1. Polaris is too cold: |
|||
Leave the machine on for a few moments and refresh. |
|||
2. Subject tracker or stylus is not seen in the field of view: |
|||
Move the subject’s chair until they are seen. |
|||
Adjust the height of the camera stand. |
|||
3. All else: |
|||
Refresh unit or restart Brainsight. |
|||
If that fails: turn off unit, wait 30 seconds, restart. |
|||
==== Registration ==== |
|||
Troubleshooting: |
|||
Holding the stylus in both hands for stability, you will mark the anatomical landmarks selected in a previous step. |
|||
<br> |
|||
''Note: It is important not to press too deeply into the participant's skin. This will distort the registration. Likewise, be sure not to move or the location will be incorrectly recorded.'' |
|||
* Click the “Speech Recognition” or “I/OBox Switch” option boxes to sample independently. |
|||
Polaris is too cold: |
|||
* Identify the landmark with the stylus |
|||
Leave the machine on for a few moments and refresh. |
|||
* Holding the stylus steady, say “SAMPLE” or step on the foot pedal. |
|||
* If you have an assistant, they can click “Sample & Go to Next Landmark” for you, and give you feedback on the camera view (green or red) for the subject tracker and stylus. |
|||
Move the subject’s chair until they are seen. |
|||
The Polaris will give you feedback as you progress through: |
|||
Adjust the height of the camera stand. |
|||
'''Successful:''' Polaris will chime and Brainsight will announce the next location to sample. |
|||
All else: |
|||
'''Unsuccessful:''' Polaris will give a negative beep. |
|||
'''Troubleshooting:''' |
|||
-Make sure both the subject tracker and stylus are in view of the camera. |
|||
-Rotate the stylus |
|||
-Ask the subject to look in a different direction. |
|||
==== Validation ==== |
|||
Refresh unit. |
|||
Check your coregistration against the anatomical data. Brainsight will tell you the distance between the landmark and the sample, and the landmark and the skin. |
|||
* Revisit the landmark samples. |
|||
If that fails: turn off unit, wait 30 seconds, restart. |
|||
* Run the stylus across the scalp. The crosshairs should follow the skull without intruding into the skull or introducing distance above. |
|||
You ideally want less than 3mm (measurement will be green), but less than 5mm(measurement will be orange) is acceptable. Otherwise the distance will be red and you will need to correct it before moving on. |
|||
'''Troubleshooting:''' |
|||
=== Registration === |
|||
* If the measurements are unacceptable, it is either because the landmarks were incorrectly set (possibly due to distortions in the MRI) or because of poor sampling. |
|||
* Resample or reset the landmarks until the validation is acceptable. When the registration is accurate, proceed to the “Electrodes” tab. |
|||
Holding the stylus in both hands for stability, mark the anatomical landmarks selected in a previous step. |
|||
Click the “Speech Recognition” or “I/OBox Switch” option boxes to sample independently. |
|||
Identify the landmark with the stylus |
|||
Holding the stylus steady, say “SAMPLE” or step on the foot pedal. |
|||
If you have an assistant, they can click “Sample & Go to Next Landmark” for you, and give you feedback on the camera view (green or red) for the subject tracker and stylus. |
|||
Polaris will give you feedback |
|||
Successful: Polaris will chime and Brainsight will announce the next location to sample. |
|||
Unsuccessful: Polaris will give a negative beep. |
|||
Make sure both the subject tracker and stylus are in view of the camera. |
|||
Rotate the stylus |
|||
Ask the subject to look in a different direction. |
|||
=== Validation === |
|||
Revisit the landmark samples. Brainsight will tell you the distance between the landmark and the sample, and the landmark and the skin. You ideally want less than 3mm (measurement will be green) but less than 5mm(measurement will be orange) is acceptable. Otherwise the distance will be red. |
|||
Run the stylus across the scalp. The crosshairs should follow the skull without intruding into the skull or introducing distance above. |
|||
If the measurements are unacceptable, it is either because the landmarks were incorrectly set (possibly due to distortions in the MRI) or because of poor sampling. |
|||
Resample or reset the landmarks until the validation is acceptable. When the registration is accurate, proceed to the “Electrodes” tab. |
|||
=== Electrodes === |
=== Electrodes === |
||
Click “Add From...” and select “Cap Layout” from the drop-down menu. In the pop-up window, select the file you wish to use. |
* Click “Add From...” and select “Cap Layout” from the drop-down menu. In the pop-up window, select the file you wish to use. For most studies, you can use “MEGCoreStandardCapLayout.txt”. This is recommended, at least as a starting point, if you use the MEG Core preprocessing pipeline. For EEG studies, you may prefer a file that includes your EEG cap layout so you can digitize your electrodes as well. |
||
Alternatively, you can create your own layout. At a minimum, you will need the following points: |
|||
For most studies, you can use “MEGCoreStandardCapLayout.txt”. This is recommended, at least as a starting point, if you use the MEG Core preprocessing pipeline – talk to Jeff Stout. |
|||
For EEG studies, you may prefer a file that includes your EEG cap layout so you can digitize your electrodes as well. |
|||
Alternatively, you can create your own layout. Minimally, for analysis you will need: |
|||
# Nasion |
# Nasion |
||
# |
|||
# Left Ear |
# Left Ear |
||
# |
|||
# Right Ear |
# Right Ear |
||
# |
|||
Holding the stylus with both hands, sample the marker coil locations and optional cloud points. Brainsight will give you feedback as before. |
* Holding the stylus with both hands, sample the marker coil locations and optional cloud points. Brainsight will give you feedback as before. |
||
* When all the electrode points are satisfactorily sampled, click “Export to File” and save under the subject’s MEG Hashcode. |
|||
;* Saving as (.txt) is recommended, and we have scripts for converting .txt to an AFNI tagset (.tag). LOCATOR file format (.elp) is supported, but requires a few additional steps. Ask the MEG Core staff if this is something you want to do. |
|||
* Save the Brainsight project under the subject’s MEG Hashcode. |
|||
== POST-REGISTRATION== |
|||
When all the electrode points are satisfactorily sampled, click “Export to File” and save under the subject’s MEG Hashcode. |
|||
Note about format: |
|||
.txt is recommended, and we have scripts for converting this to an AFNI tagset (.tag). If you plan |
|||
LOCATOR file format (.elp) is supported |
|||
Save the Brainsight project under the subject’s MEG Hashcode. |
|||
* Prep subject for entering the MSR as usual (bathroom, demetal, apply marker coils, etc.) |
|||
== Post-registration == |
|||
* Save registration date to flash drive and transfer to end destination. |
|||
* Turn off the Mac if you are finished, or lock the screen to secure it, if you plan to use it later in your session. |
|||
* Turn off the Polaris Vicra via the upper rocker switch on the control unit. |
|||
* The lower rocker switch controls power to the whole unit. If you are finished, turn off. |
|||
* '''Disinfect''' the Brainsight stylus and surfaces using a hospital approved disinfectant such as Hydrogen Peroxide, 70-90% ethyl alcohol or isopropyl alcohol after each subject/patient use. |
|||
Prep subject for entering the MSR as usual (bathroom, demetal, apply marker coils, etc.) |
|||
The stylus handle and tracker headband can be washed as usual, but special care must be taken with the reflective spheres. The spheres must be dipped in hydrogen peroxide (container provided) and allowed to air dry. |
|||
== NEXT STEPS == |
|||
Save registration date to flashdrive and transfer to end destination. |
|||
* Consult [[MEG Software and Analysis| MEG Software and Analysis]] for more information on converting the exported Brainsight .txt files to AFNI tags or MNE transforms. |
|||
You can leave the Mac on hen shut down the Vicra via the upper rocker switch, and the whole unit via the lower rocker switch. |
|||
:*bstags.py is all you need to convert .txt files to AFNI, and to continue preprocessing for SAM analysis as you would normally. |
|||
:*nih2mne pipeline will set you up for preprocessing with MNE. |
|||
Latest revision as of 10:11, 22 November 2022
OVERVIEW
Brainsight is a useful tool for modeling anatomical data for coregistration with the MEG data collected during your study. With Brainsight, you are able to set anatomical landmarks, generate a 3D rending of the head, and mark the locations of MEG marker coils and EEG electrodes. Coregistration can be done quickly before your recording and eliminates the need to do it by hand in AFNI at a later date (although, we recommend verifying your coregistration in AFNI or other similar software as you go along).
This guide is intended to be a resource for MEG Core users who have already been trained to use Brainsight in our lab. If you would like to schedule training, you can reach out to Anna Namyst directly or submit a request via the Staff Support Request form.
To learn more about Brainsight, you can read more here.
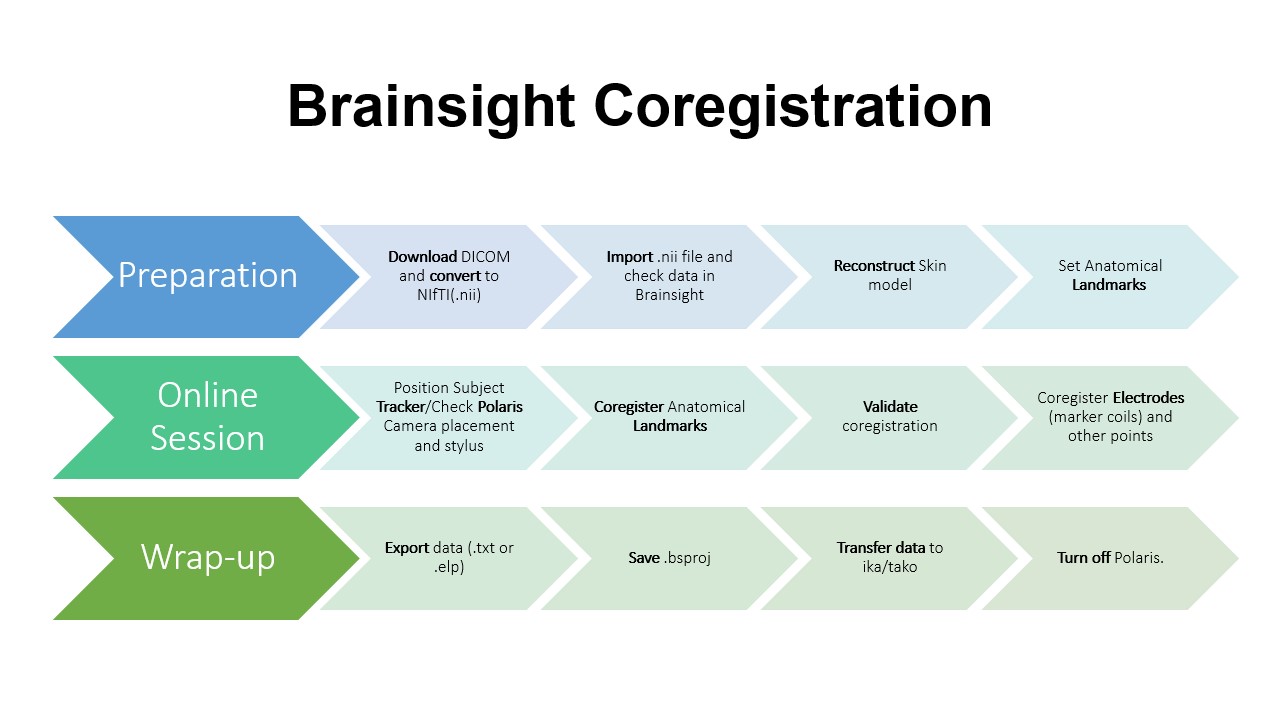
PREPARATION
Note: You will need your subject’s MRI data before your MEG appointment.
Downloading/Preparing the MRI data
If your participant’s data was collected here prior to October 2022, you can download their data from the FMRIF DICOM Archive:
https://gold.nimh.nih.gov/
If your participant’s data was collected here AFTER October 2022, you can download their data from the FMRIF XNAT Archive:
https://fmrif-xnat.nimh.nih.gov/
If you don’t have access to either data repository – talk to your PI.
Transform the DICOM to NIfTI (.nii) format
- Locate the MP RAGE
The README file from the session will tell you which directory to use. If your subject doesn’t have an MPRAGE, you can use a suitable alternative (i.e. SPGR). If you’re unsure, consult your PI or MEG Core staff.
- Navigate to that directory in terminal and use the following command:
Dimon -infile_prefix anat_t1w_mp_rage_1mm_abcd -GERT_Rico -gert_write_as_nifti –gert_create_dataset
(If you use something other than MPRAGE, replace the prefix ‘anat_t1w_mp_rage_1mm_abcd’ accordingly.)
For more information on Dimon-- https://afni.nimh.nih.gov/pub/dist/doc/program_help/Dimon.html
- Generate MEG Hashcode for your subject
If you don’t have the opportunity to do it before your study, label your file something generic (I.e. SubjectXDate.nii). You will be able to rename it later; the primary goal is to remove the PII that NMRIF includes in the filename.
PII may still be buried in the header text of the data; treat the files accordingly.
Transferring data to the Brainsight Mac
- Transfer your data to your directory on tako
- Transfer your data from tako to one of Linux computers in the MEG Lab (namako or kani)
scp -r [SOURCE]@tako.nimh.nih.gov:/tmp/[MRI data] [DESTINATION]
- Using the dedicated MEG Lab flash drive, transfer your data to the Mac and file it in the appropriate MEGLAB directory for your study
BRAINSIGHT COREGISTRATION
Importing data (May be done before Subject arrives)
- Launch the Brainsight program and select “New Empty Project”. Note: If you do not have access to your subject’s MRI, choose “New MNI Head Project” and follow the Brainsight User Manual instructions for using an average brain template (see chapter 6). This is generally not recommended for our analyses
- Select subject’s anatomical data. NIfTI files are recommended by MEG Core, but other accepted formats are MINC, Analyze, DICOM, PAR/REC, or BrainVoyager VMR.
- When the data is loaded, select “Show Image and Details” to view the metadata and image volume, or proceed to the next step.
Automatic Skin Reconstruction
- From the 3D Reconstruction tab, select “Skin” from the “New” drop-down menu.
- If needed, adjust threshold and select “keep largest piece” to isolate the head from any MR noise
Setting anatomical landmarks
Note: These are anatomical landmarks clearly identifiable from the MRI. You are not placing the marker coils at this step.
- From the “Landmarks” tab, select “Configure Landmarks”. The landmarks window will display the Skin reconstruction and coronal, sagittal, and transverse slices. The cursor cross-hairs will be displayed in all four image windows.
- With the cursor cross-hairs identify the Nasion, LPA (left preauricular), and RPA (right preauricular). Rather than using the tragus, we will use the supratrageal notch -- notch above the tragus where the tragus joins the upper ear. This point is less likely to be distorted in the MRI by earplugs or head pads.
- For additional precision, you can select a fourth point on the tip of the nose or the outer canthus. You may use whatever point as long as it is unambiguously identifiable and fixed.
- Now you are ready to start a new session. At this point, you can suspend your project or move directly to the registration session.
- If you will be using OPMs, proceed to the Targets tab and complete the target trajectory setup for the OPM rig before moving to the registration session:
Subject Preparation
Personal protective equipment must be worn from this point onward. The researcher (you) must wear gloves, and the participant must put on a skull cap before wearing the tracker headband. After the session, swab the stylus and the tracker band with alcohol.
- Draw the marker coil locations on the subject.
- 1.5cm above the nasion, 1.5cm from the tragus on a line with the outer canthus of the eye.
- Give the participant a skull cap and place the subject tracker headband on their head. For EEG subjects who have already been gelled, use a green surgical cap, but try to get it as close to the scalp as possible.
- Position them in the chair and select “New Online Session” from the session window. To select an existing session, highlight the desired session and select “Resume Session”.
Registration Session
With the Session window open, go through the following tabs(steps):
Targets (optional)
The anatomical landmarks selected can be imported here, but this section was intended for other applications (i.e. TMS, surgery). This section may be updated as we find applications for this section.
Polaris
Ensure that the Polaris Vicra is on and operational. We recommend turning on the Polaris as soon as you arrive so it has time to warm up.
Troubleshooting:
1. Polaris is too cold:
Leave the machine on for a few moments and refresh.
2. Subject tracker or stylus is not seen in the field of view:
Move the subject’s chair until they are seen.
Adjust the height of the camera stand.
3. All else:
Refresh unit or restart Brainsight.
If that fails: turn off unit, wait 30 seconds, restart.
Registration
Holding the stylus in both hands for stability, you will mark the anatomical landmarks selected in a previous step.
Note: It is important not to press too deeply into the participant's skin. This will distort the registration. Likewise, be sure not to move or the location will be incorrectly recorded.
- Click the “Speech Recognition” or “I/OBox Switch” option boxes to sample independently.
- Identify the landmark with the stylus
- Holding the stylus steady, say “SAMPLE” or step on the foot pedal.
- If you have an assistant, they can click “Sample & Go to Next Landmark” for you, and give you feedback on the camera view (green or red) for the subject tracker and stylus.
The Polaris will give you feedback as you progress through:
Successful: Polaris will chime and Brainsight will announce the next location to sample.
Unsuccessful: Polaris will give a negative beep.
Troubleshooting:
-Make sure both the subject tracker and stylus are in view of the camera.
-Rotate the stylus
-Ask the subject to look in a different direction.
Validation
Check your coregistration against the anatomical data. Brainsight will tell you the distance between the landmark and the sample, and the landmark and the skin.
- Revisit the landmark samples.
- Run the stylus across the scalp. The crosshairs should follow the skull without intruding into the skull or introducing distance above.
You ideally want less than 3mm (measurement will be green), but less than 5mm(measurement will be orange) is acceptable. Otherwise the distance will be red and you will need to correct it before moving on. Troubleshooting: * If the measurements are unacceptable, it is either because the landmarks were incorrectly set (possibly due to distortions in the MRI) or because of poor sampling. * Resample or reset the landmarks until the validation is acceptable. When the registration is accurate, proceed to the “Electrodes” tab.
Electrodes
- Click “Add From...” and select “Cap Layout” from the drop-down menu. In the pop-up window, select the file you wish to use. For most studies, you can use “MEGCoreStandardCapLayout.txt”. This is recommended, at least as a starting point, if you use the MEG Core preprocessing pipeline. For EEG studies, you may prefer a file that includes your EEG cap layout so you can digitize your electrodes as well.
Alternatively, you can create your own layout. At a minimum, you will need the following points:
- Nasion
- Left Ear
- Right Ear
- Holding the stylus with both hands, sample the marker coil locations and optional cloud points. Brainsight will give you feedback as before.
- When all the electrode points are satisfactorily sampled, click “Export to File” and save under the subject’s MEG Hashcode.
- Saving as (.txt) is recommended, and we have scripts for converting .txt to an AFNI tagset (.tag). LOCATOR file format (.elp) is supported, but requires a few additional steps. Ask the MEG Core staff if this is something you want to do.
- Save the Brainsight project under the subject’s MEG Hashcode.
POST-REGISTRATION
- Prep subject for entering the MSR as usual (bathroom, demetal, apply marker coils, etc.)
- Save registration date to flash drive and transfer to end destination.
- Turn off the Mac if you are finished, or lock the screen to secure it, if you plan to use it later in your session.
- Turn off the Polaris Vicra via the upper rocker switch on the control unit.
- The lower rocker switch controls power to the whole unit. If you are finished, turn off.
- Disinfect the Brainsight stylus and surfaces using a hospital approved disinfectant such as Hydrogen Peroxide, 70-90% ethyl alcohol or isopropyl alcohol after each subject/patient use.
The stylus handle and tracker headband can be washed as usual, but special care must be taken with the reflective spheres. The spheres must be dipped in hydrogen peroxide (container provided) and allowed to air dry.
NEXT STEPS
- Consult MEG Software and Analysis for more information on converting the exported Brainsight .txt files to AFNI tags or MNE transforms.
- bstags.py is all you need to convert .txt files to AFNI, and to continue preprocessing for SAM analysis as you would normally.
- nih2mne pipeline will set you up for preprocessing with MNE.