BIDS GUIs
What is BIDS
https://bids-specification.readthedocs.io/en/stable/
BIDS is a standard specification for neuroimaging/physiology data. This currently includes at least: MRI, fMRI, DTI, EEG, MEG, fNIRS (and possibly ECOG/sEEG). BIDS typically describes how RAW data is organized - and processed data is located in the bids_dir/derivatives/{AnalysisPackage}/{SUBJECT}/... The main advantage is that common code can be generated to process data organized in a standard format. Therefore, you should be able to import the bids data into any number of neurophysiological packages (MNE, Brainstorm, SPM, Fieldtrip, ...). Additionally, standardized processing packages known as BIDS apps can be used to process the data in the same way as long as the data is organized in BIDS.
On Biowulf
Start a compute node from the terminal
sinteractive --mem=16G --cpus-per-task=4 --gres=lscratch:50 #adjust mem and cpus accordingly
module use --append /data/MEGmodules/modulefiles module load mne/dev1.5.1
Create BIDS Dataset Using GUI
https://megcore.nih.gov/MEG/BIDS_gui_080224_anon.pdf
make_meg_bids_gui.py
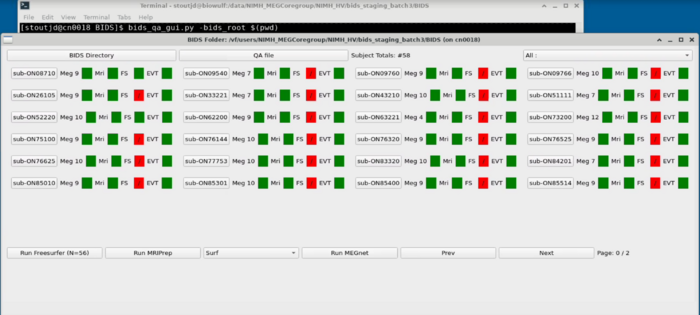
QA GUI Overview
Launch the QA gui
bids_qa_gui.py -bids_root <<PATH to BIDS dir>>
Project Panel
Subject Panel
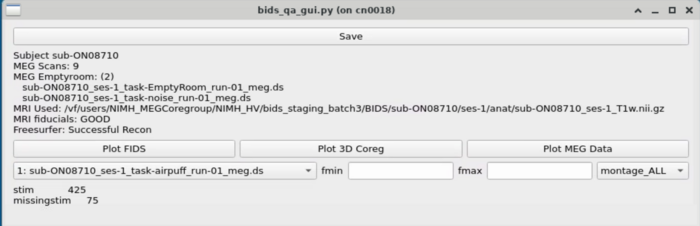
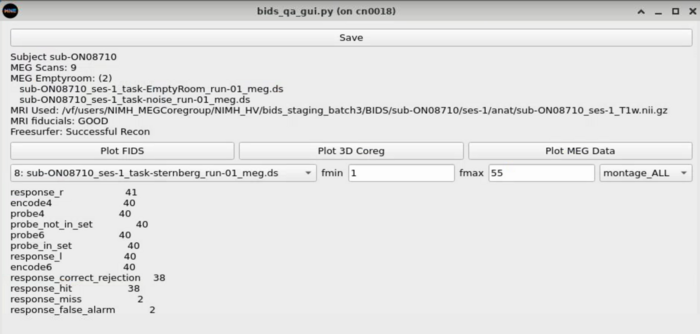
Task Switcher
Changing the task - shows the number of epochs in the selected task
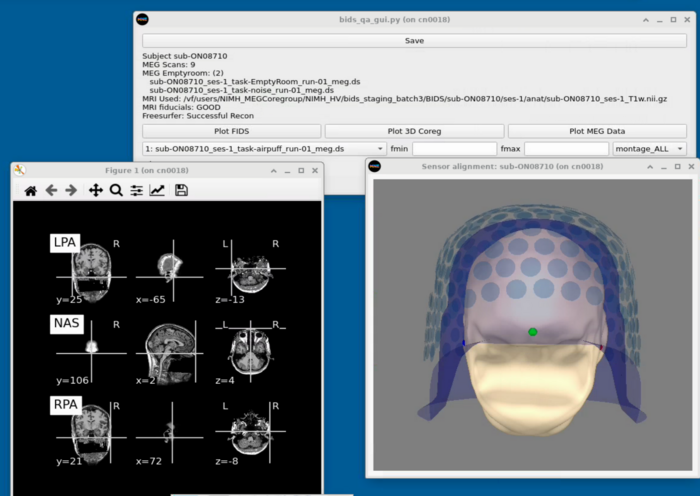
MRI Viewing
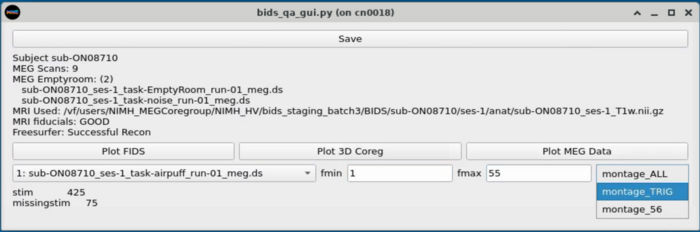
MEG Plotting
Dropdown with MEG plotting options
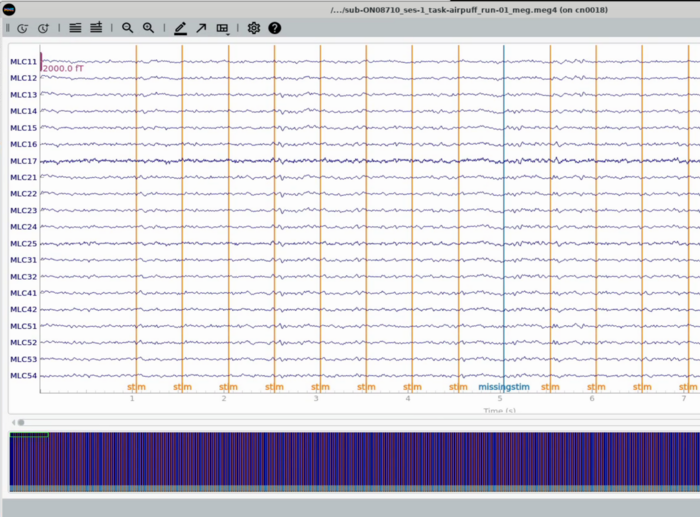
Plot All MEG channels (unorganized view of All channels)
Plot whole head - subset of meg channels representing the Left (top half) to Right (bottom half) display
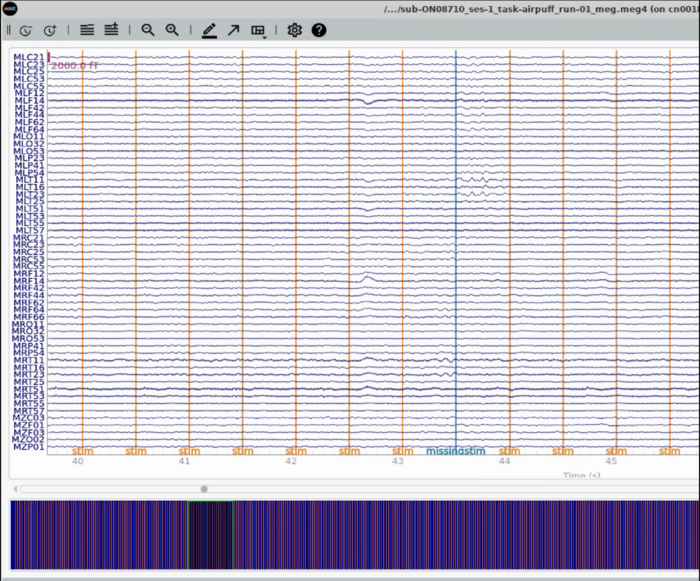
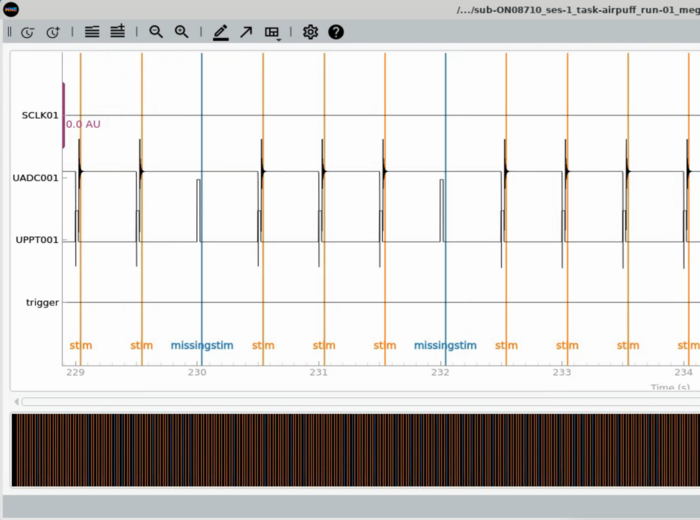
Plot Trigger Channels
Note the auditory delay added to the triggerline pulse