Analysis AWS: Difference between revisions
No edit summary |
|||
| (11 intermediate revisions by the same user not shown) | |||
| Line 4: | Line 4: | ||
== Accessing compute resources on AWS: == |
== Accessing compute resources on AWS: == |
||
Speak with Jeff Stout to get the current IP address and PEM key <br> |
Speak with Jeff Stout to get the current IP address and PEM key <br> |
||
<br> |
|||
chmod 400 AWS.pem |
|||
${PATH} is the path to the PEM key provided to you |
${PATH} is the path to the PEM key provided to you |
||
ssh -X -i |
ssh -X -i AWS.pem user1@ec2-3-230-155-59.compute-1.amazonaws.com |
||
'''Windows Laptops''' do not have X11 installed natively. Download an Xterm software for visualization. <br> |
|||
MobaXterm has been tested to work: https://mobaxterm.mobatek.net/download.html |
|||
== Jupyter Notebook Over a Remote Connection == |
== Jupyter Notebook Over a Remote Connection == |
||
| Line 23: | Line 28: | ||
== Installed software == |
== Installed software == |
||
Human Neocortical Neurosolver |
'''Human Neocortical Neurosolver ''' <br> |
||
HNN website: https://hnn.brown.edu/ |
|||
conda deactivate |
|||
hnn |
hnn |
||
#hnn is installed in the system level python >> may need to conda deactivate first |
|||
MNE python, Eelbrain, PyCTF |
'''MNE python, Eelbrain, PyCTF''' <br> |
||
MNE website: https://mne.tools/stable/index.html <br> |
|||
| ⚫ | |||
Eelbrain website: https://eelbrain.readthedocs.io/en/stable/ <br> |
|||
PyCTF website: https://megcore.nih.gov/index.php/MEG_Software_and_Analysis#MEG_Core_pyctf_tools_ported_to_Python_3 <br> |
|||
| ⚫ | |||
ipython |
|||
import eelbrain, mne, pyctf |
|||
CTF software |
'''CTF software''' |
||
singularity shell /opt/ctf/ctf.img |
singularity shell /opt/ctf/ctf.img |
||
$ctf_command |
$ctf_command |
||
| Line 36: | Line 47: | ||
singularity exec /opt/ctf/ctf.img $ctf_command |
singularity exec /opt/ctf/ctf.img $ctf_command |
||
SAM version 5 |
'''SAM version 5''' |
||
sam_cov ... |
sam_cov ... |
||
sam_3d ... |
sam_3d ... |
||
Afni |
'''Afni''' |
||
afni -dset $MRI_file |
afni -dset $MRI_file |
||
==Done== |
|||
SAM v5 |
|||
singularity ? >> CTF |
|||
Afni |
|||
Jupyter |
|||
HNN |
|||
==TODO== |
|||
Freesurfer |
|||
Reinstall MNE python |
|||
Pyctf |
|||
FSL |
|||
Brainstorm |
|||
Fieldtrip |
|||
Spyder |
|||
Latest revision as of 12:15, 7 November 2019
For Workshop members without access to computing resources - Preconfigured Amazon Web Services servers can be used
Accessing compute resources on AWS:
Speak with Jeff Stout to get the current IP address and PEM key
chmod 400 AWS.pem
${PATH} is the path to the PEM key provided to you
ssh -X -i AWS.pem user1@ec2-3-230-155-59.compute-1.amazonaws.com
Windows Laptops do not have X11 installed natively. Download an Xterm software for visualization.
MobaXterm has been tested to work: https://mobaxterm.mobatek.net/download.html
Jupyter Notebook Over a Remote Connection
1) Run one terminal to start the notebook and forward the outputs to local web browser:
ssh -i AWS.pem -L 8887:localhost:8887 ubuntu@IP_ADDRESS conda activate workshop jupyter notebook --no-browser --port=8887
2) Copy the output of the jupyter notebook command (see example below) and enter into your laptop web browser (Firefox, Chrome, etc.). Do not click on the link - it will try to launch on the AWS computer
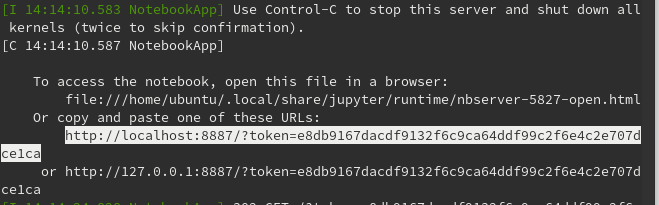
3) Use the jupyter notebook to analyze MEG data with python tools
Installed software
Human Neocortical Neurosolver
HNN website: https://hnn.brown.edu/
conda deactivate hnn
MNE python, Eelbrain, PyCTF
MNE website: https://mne.tools/stable/index.html
Eelbrain website: https://eelbrain.readthedocs.io/en/stable/
PyCTF website: https://megcore.nih.gov/index.php/MEG_Software_and_Analysis#MEG_Core_pyctf_tools_ported_to_Python_3
conda activate workshop # This conda environment has the MNE, Eelbrain, and PyCtf toolboxes installed ipython import eelbrain, mne, pyctf
CTF software
singularity shell /opt/ctf/ctf.img $ctf_command
singularity exec /opt/ctf/ctf.img $ctf_command
SAM version 5
sam_cov ... sam_3d ...
Afni
afni -dset $MRI_file